| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,416,582 – 19,416,693 |
| Length | 111 |
| Max. P | 0.945620 |

| Location | 19,416,582 – 19,416,693 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
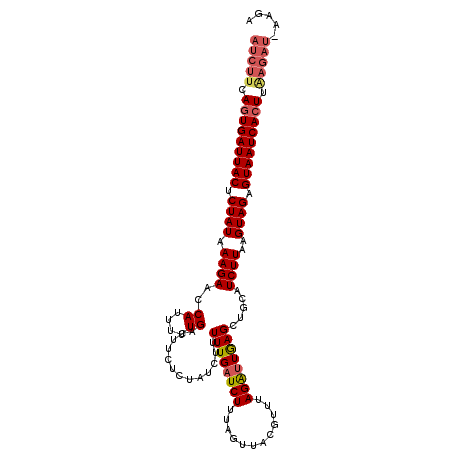
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.24 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19416582 111 + 22407834 UCUUCAUCUGAAGUGAUUACUCUACUUAAGAUGCACCUUAAUCUU-CCGUAUCUAAAGAUCAAAAAGAUAGAGAAGUCAAAAAUGGUUCUUUAUAGCGUAAUCACUGAAGAU .....((((..(((((((((.((.(((.((((((...........-..)))))).))).........((((((((.(((....))))))))))))).)))))))))..)))) ( -29.82) >DroSec_CAF1 19475 112 + 1 UCUUUAUCUCAACUGAUUACACUACUUAAGAUGCAGCUCAUCCUAAACGUAACUAAAGACCAAAAAGAUAGAGAAGUCAAAAAUGGUUCUUUAUAGAGUAAUCACUGAAGAU .....((((((..(((((((.(((.(((.((((.....)))).))).......(((((((((....(((......))).....))).))))))))).))))))).)).)))) ( -19.90) >DroSim_CAF1 20071 104 + 1 U--------UAAGUGAUUACUCUACUUAAGAUGCAGCUCAAUCUAAACGUAACUAAAGAUCAAAAAGAUAGAGAAGUCAAAAGUGGUUCUUUAUAGAGUAAUCACUGAAGAU .--------..(((((((((((((....((((........)))).........(((((((((....(((......))).....))).)))))))))))))))))))...... ( -25.70) >consensus UCUU_AUCUCAAGUGAUUACUCUACUUAAGAUGCAGCUCAAUCUAAACGUAACUAAAGAUCAAAAAGAUAGAGAAGUCAAAAAUGGUUCUUUAUAGAGUAAUCACUGAAGAU ...........(((((((((.((.(((.((.(((..............))).)).))).........((((((((.(((....))))))))))))).)))))))))...... (-18.91 = -19.24 + 0.33)

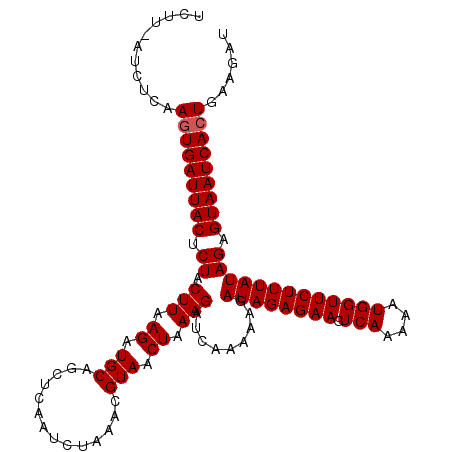
| Location | 19,416,582 – 19,416,693 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -16.45 |
| Energy contribution | -18.23 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19416582 111 - 22407834 AUCUUCAGUGAUUACGCUAUAAAGAACCAUUUUUGACUUCUCUAUCUUUUUGAUCUUUAGAUACGG-AAGAUUAAGGUGCAUCUUAAGUAGAGUAAUCACUUCAGAUGAAGA ((((..(((((((((.((((.((((..((((((...(((((.(((((...........))))).))-)))...))))))..))))..)))).)))))))))..))))..... ( -31.60) >DroSec_CAF1 19475 112 - 1 AUCUUCAGUGAUUACUCUAUAAAGAACCAUUUUUGACUUCUCUAUCUUUUUGGUCUUUAGUUACGUUUAGGAUGAGCUGCAUCUUAAGUAGUGUAAUCAGUUGAGAUAAAGA (((((.(.(((((((.(((((((((.(((.....((........))....)))))))))......(((((((((.....)))))))))))).))))))).).)))))..... ( -29.00) >DroSim_CAF1 20071 104 - 1 AUCUUCAGUGAUUACUCUAUAAAGAACCACUUUUGACUUCUCUAUCUUUUUGAUCUUUAGUUACGUUUAGAUUGAGCUGCAUCUUAAGUAGAGUAAUCACUUA--------A ......((((((((((((((.((((........(((((.....(((.....)))....))))).((((.....))))....))))..))))))))))))))..--------. ( -24.90) >consensus AUCUUCAGUGAUUACUCUAUAAAGAACCAUUUUUGACUUCUCUAUCUUUUUGAUCUUUAGUUACGUUUAGAUUGAGCUGCAUCUUAAGUAGAGUAAUCACUUAAGAU_AAGA (((((.(((((((((.((((.((((..((....)).............((((((((............)))))))).....))))..)))).))))))))).)))))..... (-16.45 = -18.23 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:38 2006