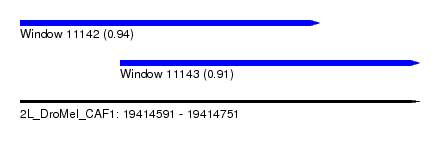
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,414,591 – 19,414,751 |
| Length | 160 |
| Max. P | 0.935690 |

| Location | 19,414,591 – 19,414,711 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -31.68 |
| Energy contribution | -30.72 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19414591 120 + 22407834 AUCGCUCCGAGCACUGGACCUCCGGCAAUGAUUUGGGCAAAACCGGCACCCACUACUGGUUCUCUAAUGCACAACUCGUGACCAUUAAGCGUUGGGCGCCCAAACAACCAGACAAUGCUG .........((((((((....))))...((.(((((((..((((((.........)))))).((((((((..................)))))))).))))))))).........)))). ( -31.17) >DroSec_CAF1 17550 120 + 1 AUCGCUCCGAGCACUGGACCUCCGGCAAUGACUUGGGCAAAACCGGCACCCACUACUGGUUCUCCAACGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAAGCAACCAGACAAUGCUG ...((.(((((((((((....))))...)).))))))).....(((((.......((((((((....((((((((.(.((.....)).).))))))))....)).))))))....))))) ( -42.20) >DroSim_CAF1 17803 120 + 1 AUCGCUCCGAGCACUGGACCUCCGGCAAUGACUUGGGCAAAACCGGCACCCACUACUGGUUCUCCAAUGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAAGCAACCAGACAAUGCUG .........((((((((....))))...((.(((((((..((((((.........)))))).......(((((((.(.((.....)).).)))))))))))))))).........)))). ( -41.80) >DroEre_CAF1 17137 120 + 1 AUCGCUCCGAGCACUGGACUUCCGGCAAUGACUUGGGCCAAACCGGCACCCACAAUUGGUUCUCCAAUGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAACCAACCAGACAACUCUG ...((.(((((((((((....))))...)).)))))))......(((.......(((((....)))))(((((((.(.((.....)).).))))))))))........((((....)))) ( -38.30) >DroYak_CAF1 17850 120 + 1 AUCGCUCUGAGCACUGGACUUCUGGCAAUGACUUGGCCCGAACCGGAGACUACUAUUGGUUCUCCAAUGUCCAACUCGUGACCAUUAAGCGUUGGGCGCCCAACCAACCAGACAACUCUG ...(((.((..(((((((((((.(((.........))).)))..((((((((....))).)))))...)))))....)))..))...)))(((((....)))))....((((....)))) ( -33.90) >consensus AUCGCUCCGAGCACUGGACCUCCGGCAAUGACUUGGGCAAAACCGGCACCCACUACUGGUUCUCCAAUGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAACCAACCAGACAAUGCUG ...((.(((((((((((....))))...)).))))))).....(((((.......((((((......((((((((.(.((.....)).).)))))))).......))))))....))))) (-31.68 = -30.72 + -0.96)

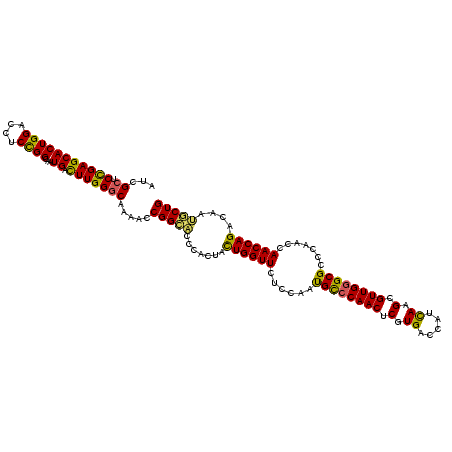
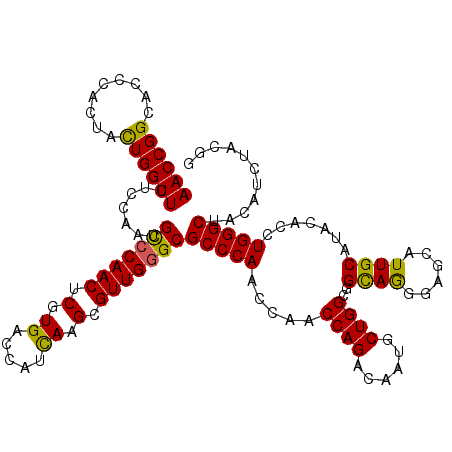
| Location | 19,414,631 – 19,414,751 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -32.78 |
| Energy contribution | -31.94 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19414631 120 + 22407834 AACCGGCACCCACUACUGGUUCUCUAAUGCACAACUCGUGACCAUUAAGCGUUGGGCGCCCAAACAACCAGACAAUGCUGGCGGUAGGGAGCAUUGCAUACACUUGGGCUACAUCUACGG ..(((((..(((....)))....(((((((..................)))))))))((((((.......(.(((((((..(....)..))))))))......))))))........))) ( -29.79) >DroSec_CAF1 17590 120 + 1 AACCGGCACCCACUACUGGUUCUCCAACGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAAGCAACCAGACAAUGCUGGCGGCAGGGAGCAUUGCAUACACCUGGGCUACAUCUACAA ....(...((((...((((((((....((((((((.(.((.....)).).))))))))....)).)))))).(((((((..(....)..)))))))........))))...)........ ( -38.40) >DroSim_CAF1 17843 120 + 1 AACCGGCACCCACUACUGGUUCUCCAAUGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAAGCAACCAGACAAUGCUGGCGGCAGGGAGCAUUGCAUACACCUGGGCUACAUCUACAA ((((((.........)))))).......(((((((.(.((.....)).).)))))))((((..((..((((......))))..))(((..............)))))))........... ( -38.04) >DroEre_CAF1 17177 120 + 1 AACCGGCACCCACAAUUGGUUCUCCAAUGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAACCAACCAGACAACUCUGGGGGCAAGGAGCAUUGCAUUCACUUGGGCUACAUCUACGG ..(((.........(((((....)))))(((((((.(.((.....)).).)))))))((((((((..(((((....)))))))((((......))))......))))))........))) ( -41.80) >DroYak_CAF1 17890 120 + 1 AACCGGAGACUACUAUUGGUUCUCCAAUGUCCAACUCGUGACCAUUAAGCGUUGGGCGCCCAACCAACCAGACAACUCUGGGGGCAAUGAGCAUUGCAUACACAUGGGCUACAUCUACGG ..((((((((((....))).)))))...(((((((.(...........).)))))))(((((.((..(((((....)))))))(((((....))))).......))))).........)) ( -40.10) >consensus AACCGGCACCCACUACUGGUUCUCCAAUGCCCAACUCGUGACCAUCAAGCGUUGGGCGCCCAACCAACCAGACAAUGCUGGCGGCAGGGAGCAUUGCAUACACCUGGGCUACAUCUACGG ((((((.........)))))).......(((((((.(.((.....)).).)))))))(((((.....((((......))))..((((......)))).......)))))........... (-32.78 = -31.94 + -0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:36 2006