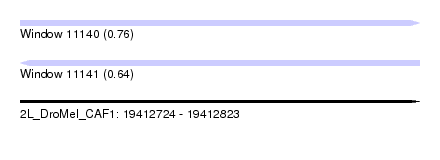
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,412,724 – 19,412,823 |
| Length | 99 |
| Max. P | 0.760300 |

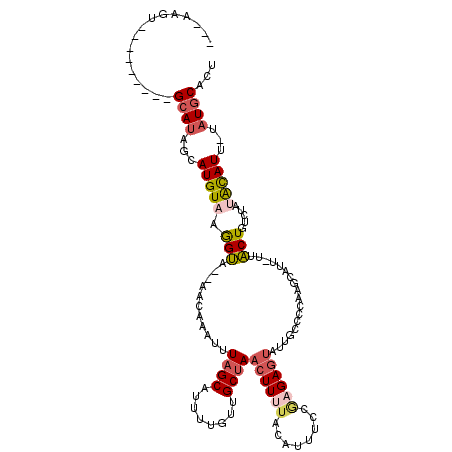
| Location | 19,412,724 – 19,412,823 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -6.70 |
| Energy contribution | -6.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
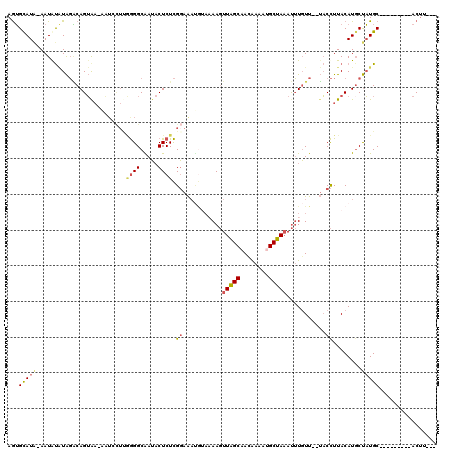
>2L_DroMel_CAF1 19412724 99 + 22407834 AGUGCAUA-AAUAUGUAGACAGUGC-G-----AGGGGCAAUACUCUCGGAAAUGUAAAAGUUAGCAAUAAAAAGCUAAAUUUGUU--UACCUUACAUGCUAUGC---------ACUU--- ((((((((-..(((((((......(-(-----((((......)))))).....(((((..(((((........))))).....))--))).))))))).)))))---------))).--- ( -30.00) >DroSec_CAF1 15656 104 + 1 AGUGCAUA-AAUGUAUAGACAGUGA-AAUCCUCGGGGCAAUACUCUCGGAAAUGUAAAAGUUAGCAACAAAAUGCUAAAUUUGUU--UACCUUACAUGCUAUGC---------ACUU--- ((((((((-.((((((((((((...-..(((..(((......)))..)))..........((((((......))))))..)))))--))...)))))..)))))---------))).--- ( -28.80) >DroSim_CAF1 15918 104 + 1 AGUGCAUA-AAUGUAUAGACAGUAA-AAUCCUCGGGGCAAUACUCUCGGAAAUGUAAAAGUUAGCAACAAAAUGCUAAAUUUGUU--UACCUUACAUGCUAUGC---------ACUU--- ((((((((-.((((((((((((...-..(((..(((......)))..)))..........((((((......))))))..)))))--))...)))))..)))))---------))).--- ( -28.80) >DroEre_CAF1 15139 107 + 1 AGUGCAUG-AAUAUAUAGGCAGUAA-GAUGCUUAGGGCAAUACUCUCGGAA------AAGUUAGCAACAAAGUGCUACAUUUGUU--UACCUUACAUCCUAUGCAUAGGAUCCACUU--- .(((((((-......((((((....-..))))))(((.....)))..(((.------..((.((((......))))))...(((.--......)))))))))))))...........--- ( -20.10) >DroYak_CAF1 15973 105 + 1 AGUGCAUG-AAUGUAUAGGCAGUAAAAAUGCUUAGGGCGAUACUCUCGGAAAUGGAUAAGUUAGCAACAAAAUGCUAAAUUUGUU--UACCUUACAUGCUAUGC---------ACUU--- ((((((((-.(((((((((((.......))))))((.(((.....)))....((((((((((((((......)))).))))))))--)))).)))))..)))))---------))).--- ( -31.40) >DroAna_CAF1 14016 110 + 1 AAAGUAUCUAAUACCUGUUCAGAAA-CUUAUUUCAGGCGAGCCUCUUGCAGACAUAAAACUUGGCAAAGAAAAGCUAAACAUCUUGUAUCUUUACAUGCCUUGC---------ACAUCAC .((((.((((((....))).))).)-))).....((((((((.((((((.((........)).)).))))...)))........((((....)))))))))...---------....... ( -15.30) >consensus AGUGCAUA_AAUAUAUAGACAGUAA_AAUCCUUGGGGCAAUACUCUCGGAAAUGUAAAAGUUAGCAACAAAAUGCUAAAUUUGUU__UACCUUACAUGCUAUGC_________ACUU___ ...(((((.........................((((.....)))).((...........(((((........)))))...........))........)))))................ ( -6.70 = -6.95 + 0.25)

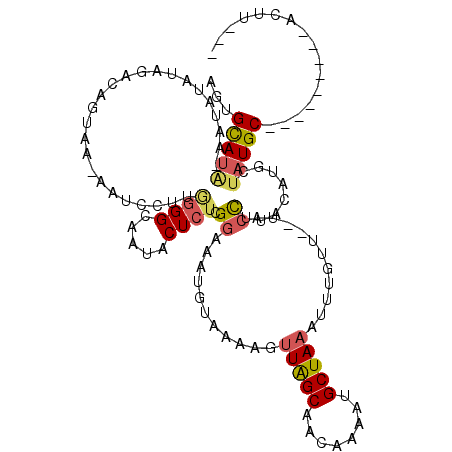
| Location | 19,412,724 – 19,412,823 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.51 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -7.61 |
| Energy contribution | -8.37 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19412724 99 - 22407834 ---AAGU---------GCAUAGCAUGUAAGGUA--AACAAAUUUAGCUUUUUAUUGCUAACUUUUACAUUUCCGAGAGUAUUGCCCCU-----C-GCACUGUCUACAUAUU-UAUGCACU ---.(((---------((((((.(((((..(((--((.....(((((........)))))..)))))......((.(((...((....-----.-))))).)))))))..)-)))))))) ( -23.10) >DroSec_CAF1 15656 104 - 1 ---AAGU---------GCAUAGCAUGUAAGGUA--AACAAAUUUAGCAUUUUGUUGCUAACUUUUACAUUUCCGAGAGUAUUGCCCCGAGGAUU-UCACUGUCUAUACAUU-UAUGCACU ---.(((---------((((((.(((((.((((--(.......(((((......)))))((((((........)))))).)))))....((((.-.....)))).))))))-)))))))) ( -29.20) >DroSim_CAF1 15918 104 - 1 ---AAGU---------GCAUAGCAUGUAAGGUA--AACAAAUUUAGCAUUUUGUUGCUAACUUUUACAUUUCCGAGAGUAUUGCCCCGAGGAUU-UUACUGUCUAUACAUU-UAUGCACU ---.(((---------((((((.(((((..(((--((.....((((((......))))))..)))))......((.((((...((....))...-.)))).))..))))))-)))))))) ( -29.30) >DroEre_CAF1 15139 107 - 1 ---AAGUGGAUCCUAUGCAUAGGAUGUAAGGUA--AACAAAUGUAGCACUUUGUUGCUAACUU------UUCCGAGAGUAUUGCCCUAAGCAUC-UUACUGCCUAUAUAUU-CAUGCACU ---..((((((..((.(((((((((((..((((--(.......(((((......)))))((((------(....))))).)))))....)))))-))).))).))...)))-)))..... ( -25.10) >DroYak_CAF1 15973 105 - 1 ---AAGU---------GCAUAGCAUGUAAGGUA--AACAAAUUUAGCAUUUUGUUGCUAACUUAUCCAUUUCCGAGAGUAUCGCCCUAAGCAUUUUUACUGCCUAUACAUU-CAUGCACU ---.(((---------((((.(.((((((((((--.......((((((......))))))............(((.....)))................))))).))))).-)))))))) ( -25.20) >DroAna_CAF1 14016 110 - 1 GUGAUGU---------GCAAGGCAUGUAAAGAUACAAGAUGUUUAGCUUUUCUUUGCCAAGUUUUAUGUCUGCAAGAGGCUCGCCUGAAAUAAG-UUUCUGAACAGGUAUUAGAUACUUU ....(((---------...((((..(((((((...(((........))).)))))))..((((((.((....)).)))))).))))...)))((-(.(((((.......))))).))).. ( -21.70) >consensus ___AAGU_________GCAUAGCAUGUAAGGUA__AACAAAUUUAGCAUUUUGUUGCUAACUUUUACAUUUCCGAGAGUAUUGCCCCAAGCAUU_UUACUGUCUAUACAUU_UAUGCACU ................((((...(((((.(((...........((((........))))((((((........))))))..................))).....)))))...))))... ( -7.61 = -8.37 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:34 2006