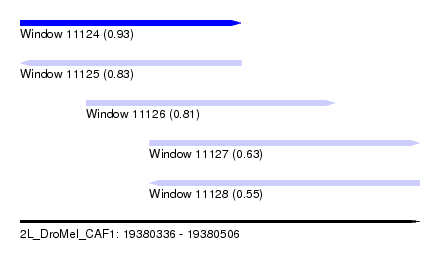
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,380,336 – 19,380,506 |
| Length | 170 |
| Max. P | 0.928078 |

| Location | 19,380,336 – 19,380,430 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -18.66 |
| Energy contribution | -21.10 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19380336 94 + 22407834 UGCCAAACUGAGAGUUAAAGGCGGCAUUG-CUGCCGAAGUUGGUGGCGGCAUUGCUGC-CGAAGUUGGUGGCGGCAUUGCUGCCGAAGUUGGAGGC .(((.(((.....)))...(((((((.((-((((((.......(((((((...)))))-)).......)))))))).))))))).........))) ( -44.44) >DroSim_CAF1 14200 94 + 1 UGCCAAACUGAGAGUUGAAGGCGGCAUUG-CUGCCGAAGUUGGAGGCGGUGUUGCUGC-CGAAGUUGGUGGCGGCAUUGCUGCCGAAGGUGGAGGC .(((..(((..........((((((...)-)))))....((((.(((((((((((..(-((....)))..))))))))))).)))).)))...))) ( -42.70) >DroWil_CAF1 24311 88 + 1 UGCCGAAUUGAGAGCCAA-------AUUGACUACCGAA-UUGACUGCCGAAUUGCUGAUUGACUUUAAAGGCAGAGUUGGUGCCGAAAUUAGAGGC .(((.((((..(.(((((-------.(((.....))).-....(((((.((...(.....)...))...)))))..)))))..)..))))...))) ( -18.60) >DroYak_CAF1 14424 94 + 1 UGCCGAGCUGAGAGUUGAAGGCGGCAUUG-CUACCGAAGUUGGAGGCGGUAUUACUGC-CAAAGUUGGAGGCGACAUUGCUGCCAAAAGUGGAGGC .(((.(((.....)))...((((((((((-((.((((..((...(((((.....))))-)))..)))).)))))...))))))).........))) ( -33.70) >consensus UGCCAAACUGAGAGUUAAAGGCGGCAUUG_CUACCGAAGUUGGAGGCGGUAUUGCUGC_CGAAGUUGGAGGCGGCAUUGCUGCCGAAAGUGGAGGC .(((.(((.....)))...((((((((((.(((((((..(((...((((.....)))).)))..))))))))))...))))))).........))) (-18.66 = -21.10 + 2.44)

| Location | 19,380,336 – 19,380,430 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -13.11 |
| Energy contribution | -13.93 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19380336 94 - 22407834 GCCUCCAACUUCGGCAGCAAUGCCGCCACCAACUUCG-GCAGCAAUGCCGCCACCAACUUCGGCAG-CAAUGCCGCCUUUAACUCUCAGUUUGGCA (((...((((..(((.(((.(((.(((.........)-)).))).))).)))........(((((.-...)))))............)))).))). ( -33.90) >DroSim_CAF1 14200 94 - 1 GCCUCCACCUUCGGCAGCAAUGCCGCCACCAACUUCG-GCAGCAACACCGCCUCCAACUUCGGCAG-CAAUGCCGCCUUCAACUCUCAGUUUGGCA ............((((....))))((((..((((..(-(..((......))..)).....(((((.-...)))))............)))))))). ( -27.70) >DroWil_CAF1 24311 88 - 1 GCCUCUAAUUUCGGCACCAACUCUGCCUUUAAAGUCAAUCAGCAAUUCGGCAGUCAA-UUCGGUAGUCAAU-------UUGGCUCUCAAUUCGGCA (((.........((((.......))))......((......)).....))).(((((-((.((.((((...-------..)))))).)))).))). ( -17.50) >DroYak_CAF1 14424 94 - 1 GCCUCCACUUUUGGCAGCAAUGUCGCCUCCAACUUUG-GCAGUAAUACCGCCUCCAACUUCGGUAG-CAAUGCCGCCUUCAACUCUCAGCUCGGCA (((.........(((.(((.(((..((.........(-((.(.....).))).........))..)-)).))).))).....(.....)...))). ( -23.17) >consensus GCCUCCAACUUCGGCAGCAAUGCCGCCACCAACUUCG_GCAGCAAUACCGCCUCCAACUUCGGCAG_CAAUGCCGCCUUCAACUCUCAGUUCGGCA (((........(((((....)))))...................................(((((.....))))).................))). (-13.11 = -13.93 + 0.81)

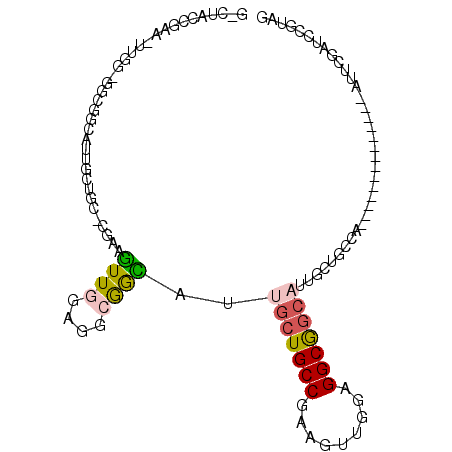
| Location | 19,380,364 – 19,380,470 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 57.19 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19380364 106 + 22407834 G-CUGCCGAAGUUGGUGGCGGCAUUGCUGC-CGAAGUUGGUGGCGGCAUUGCUGCCGAAGUUGGAGGCAGCAUUGCUGCCAAAGGUGGAGGCGGCAUUCGAUCCGUAA (-(((((........((((((((.((((((-(........(((((((...)))))))........))))))).))))))))........))))))............. ( -54.58) >DroSec_CAF1 13995 86 + 1 ---------------------CAUUGCUGC-CGAAGUUGGAGGCGGUAUUGCUGCCGAAGUUCGUGGCGGCAUUGCUGCCAAAGGUGGAGGCGGCAUUCGAUCCGUAG ---------------------...((((((-(..(.((...(((((((.((((((((.......)))))))).)))))))..)).)...)))))))............ ( -38.30) >DroSim_CAF1 14228 82 + 1 G-CUGCCGAAGUUGGAGGCGGUGUUGCUGC-CGAAGUUGGUGGCGGCAUUGCUGCCGAAGGUGGAGGCGGC------------------------AUUCGAUCCGUAG (-(((((....((((.(((((((((((..(-((....)))..))))))))))).)))).......))))))------------------------............. ( -40.10) >DroEre_CAF1 14139 82 + 1 G-CUA------------------------C-CGAAGUUCGAGGCGGCAUUGCUGCCAAAGUUGCUGGCGGCAUUGCUGCCAAAGGAGGAGGCGGCAUUCGAUCCGUAG (-(..------------------------(-(....(((..(((((((.((((((((.......)))))))).)))))))...)))...))..))............. ( -36.20) >DroWil_CAF1 24332 83 + 1 GACUACCGAA-UUGACUGCCGAAUUGCUGAUUGACUUUAAAGGCAGAGUUGGUGCCGAAAUUAGAGGCAGACUGGCCCCCA------------------------UAG ..........-....(((((.((((.(.(((..(((((......)))))..)).).).))))...)))))...........------------------------... ( -16.70) >DroYak_CAF1 14452 82 + 1 G-CUACCGAAGUUGGAGGCGGUAUUACUGC-CAAAGUUGGAGGCGACAUUGCUGCCAAAAGUGGAGGCGGCAUUCG------------------------AUCCGUAG .-((((..........(((((.....))))-)......(((..(((...(((((((.........))))))).)))------------------------.))))))) ( -29.10) >consensus G_CUACCGAA_UUGG_GGCGGCAUUGCUGC_CGAAGUUGGAGGCGGCAUUGCUGCCGAAGUUGGAGGCGGCAUUGCUGCCA______________AUUCGAUCCGUAG ...................................((((....))))..(((((((.........))))))).................................... ( -9.62 = -9.73 + 0.11)


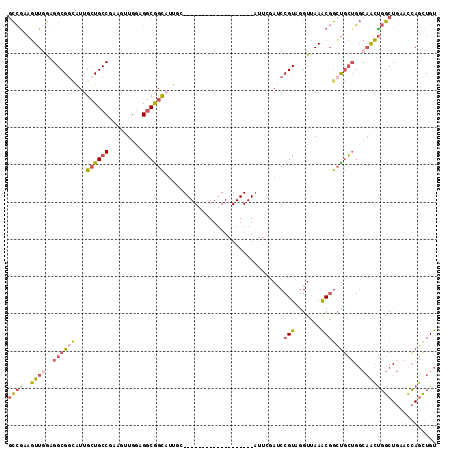
| Location | 19,380,391 – 19,380,506 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.91 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -20.49 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19380391 115 + 22407834 GCCGAAGUUGGUGGCGGCAUUGCUGCCGAAGUUGGAGGCAGCAUUGCUGCCAAAGGUGGAGGCGGCAUUCGAUCCGUAAGUCAAACGGCUGCUGGCAACUGGCUGAACCAGCUGU ((((.(((((.((((((((.(((((((.........))))))).))))))))....(((..((((........))))...)))..)))))..))))....(((((...))))).. ( -50.80) >DroSec_CAF1 14002 115 + 1 GCCGAAGUUGGAGGCGGUAUUGCUGCCGAAGUUCGUGGCGGCAUUGCUGCCAAAGGUGGAGGCGGCAUUCGAUCCGUAGGUUAAACGGCUGCUGGCAACUGGCUGAACCAGUUGU ((((...((((..((((...(((((((........(((((((...)))))))........)))))))......))))...)))).)))).....((((((((.....)))))))) ( -48.69) >DroSim_CAF1 14255 91 + 1 GCCGAAGUUGGUGGCGGCAUUGCUGCCGAAGGUGGAGGCGGC------------------------AUUCGAUCCGUAGGUUAAACGGCUGCUGGCAACUGGCUGAACCAGCUGU .....(((((.(((((((..(((((((.........))))))------------------------).......(((.......)))))))))).)))))(((((...))))).. ( -35.00) >DroEre_CAF1 14143 114 + 1 -CCGAAGUUCGAGGCGGCAUUGCUGCCAAAGUUGCUGGCGGCAUUGCUGCCAAAGGAGGAGGCGGCAUUCGAUCCGUAGGUUAAGCGGCUGCUGGCAGCUGGCUGAAUCAGUUGU -.........((..((((...(((((((.(((((((.((((...(((((((.........)))))))......))))......)))))))..)))))))..))))..))...... ( -48.80) >DroYak_CAF1 14479 91 + 1 GCCAAAGUUGGAGGCGACAUUGCUGCCAAAAGUGGAGGCGGCAUUCG------------------------AUCCGUAGGUCAAACGGCUGCUGGCAACUGGCUGAACCAGCUGU ((((.....(((..(((...(((((((.........))))))).)))------------------------.)))((((.((....))))))))))....(((((...))))).. ( -32.30) >DroMoj_CAF1 19319 88 + 1 GUUGACCUUAAAGGCGGAGUUGGUGCCGAAGCUGGAGCCGGUCUUCG------------------------AGCCGUAGGUGAAGCUAGCCGAG---GGCUGCUGGACGAUCUGU (((.((((....((((((((((((.(((....))).))))).)))).------------------------.)))..))))..(((.((((...---))))))).)))....... ( -28.60) >consensus GCCGAAGUUGGAGGCGGCAUUGCUGCCGAAGUUGGAGGCGGCAUUGC___________________AUUCGAUCCGUAGGUUAAACGGCUGCUGGCAACUGGCUGAACCAGCUGU ((((..((((..((((((...((((((.........))))))................................(((.......)))))))))..))))))))............ (-20.49 = -21.17 + 0.67)

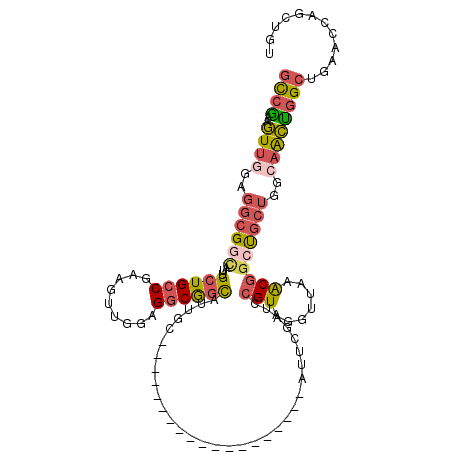
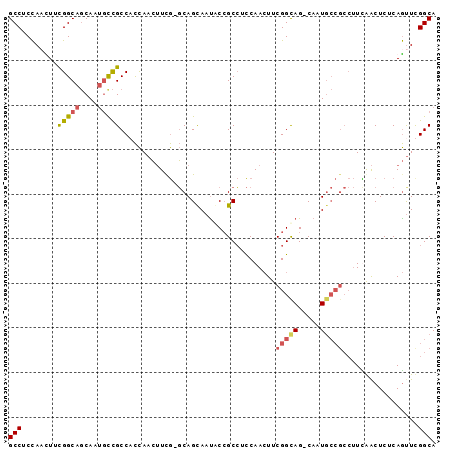
| Location | 19,380,391 – 19,380,506 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.91 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -16.32 |
| Energy contribution | -17.80 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19380391 115 - 22407834 ACAGCUGGUUCAGCCAGUUGCCAGCAGCCGUUUGACUUACGGAUCGAAUGCCGCCUCCACCUUUGGCAGCAAUGCUGCCUCCAACUUCGGCAGCAAUGCCGCCACCAACUUCGGC .(((((((.....)))))))......((((.(((......(((.((.....))..))).....((((.(((.(((((((.........))))))).))).)))).)))...)))) ( -43.40) >DroSec_CAF1 14002 115 - 1 ACAACUGGUUCAGCCAGUUGCCAGCAGCCGUUUAACCUACGGAUCGAAUGCCGCCUCCACCUUUGGCAGCAAUGCCGCCACGAACUUCGGCAGCAAUACCGCCUCCAACUUCGGC .(((((((.....)))))))......((((.........(((......(((.(((.........))).))).(((.(((.........))).)))...)))..........)))) ( -34.71) >DroSim_CAF1 14255 91 - 1 ACAGCUGGUUCAGCCAGUUGCCAGCAGCCGUUUAACCUACGGAUCGAAU------------------------GCCGCCUCCACCUUCGGCAGCAAUGCCGCCACCAACUUCGGC .(((((((.....))))))).......((((.......)))).......------------------------((((..........(((((....)))))..........)))) ( -28.35) >DroEre_CAF1 14143 114 - 1 ACAACUGAUUCAGCCAGCUGCCAGCAGCCGCUUAACCUACGGAUCGAAUGCCGCCUCCUCCUUUGGCAGCAAUGCCGCCAGCAACUUUGGCAGCAAUGCCGCCUCGAACUUCGG- ....((((((((((..((((....)))).)))........(((.((.....))..)))......(((.(((.(((.(((((.....))))).))).))).)))..)))..))))- ( -33.40) >DroYak_CAF1 14479 91 - 1 ACAGCUGGUUCAGCCAGUUGCCAGCAGCCGUUUGACCUACGGAU------------------------CGAAUGCCGCCUCCACUUUUGGCAGCAAUGUCGCCUCCAACUUUGGC .(((((((.....)))))))...((.(.(((((((.(....).)------------------------)))))).)(((.........))).))......(((.........))) ( -27.60) >DroMoj_CAF1 19319 88 - 1 ACAGAUCGUCCAGCAGCC---CUCGGCUAGCUUCACCUACGGCU------------------------CGAAGACCGGCUCCAGCUUCGGCACCAACUCCGCCUUUAAGGUCAAC ...((((((..(((((((---...)))).)))..))....((((------------------------((.....))).....((....)).........))).....))))... ( -21.40) >consensus ACAGCUGGUUCAGCCAGUUGCCAGCAGCCGUUUAACCUACGGAUCGAAU___________________GCAAUGCCGCCUCCAACUUCGGCAGCAAUGCCGCCUCCAACUUCGGC .(((((((.....)))))))......((((.........(((......(((((..........)))))......)))..........(((((....)))))..........)))) (-16.32 = -17.80 + 1.48)

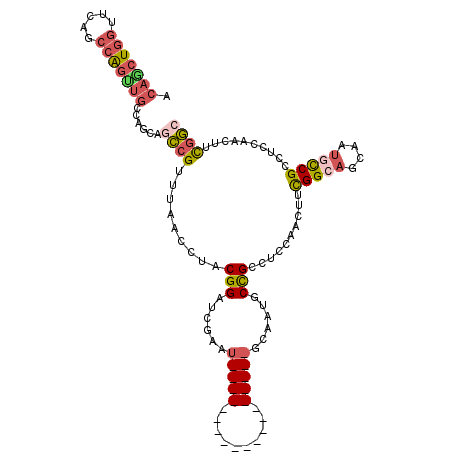
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:23 2006