
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,365,088 – 19,365,217 |
| Length | 129 |
| Max. P | 0.876786 |

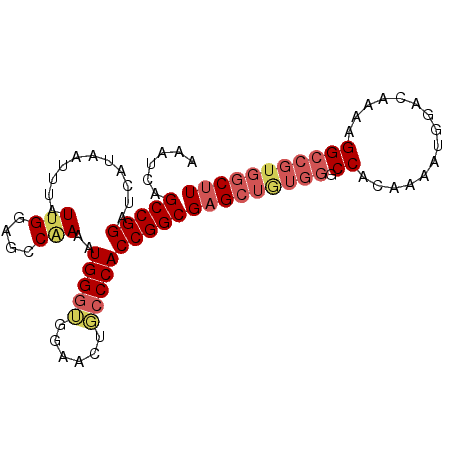
| Location | 19,365,088 – 19,365,181 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.94 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19365088 93 - 22407834 AAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGAUGGUACUGCCCACCGGCGAGCUAUGGGCCACAAAAUGGACAAAAGGCCGUGGCUU ......(((((......(((((((........)))))))((.....))..)))))(((((((((.((...............))))))))))) ( -27.46) >DroSec_CAF1 28571 87 - 1 AAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGGUGGAACUGCCCACCGGCGAGCUGUGGGCCACAAAAUGGACAAAAGG------CUU ......(((((...........(((....)))..(((((......))))))))))(((((.((..(((.....))).))...))------))) ( -27.40) >DroSim_CAF1 28961 93 - 1 AAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGGUGGAACUGCCCACCGGCGAGCUGUGGGCCACAAAAUGGACAAAAGGCCGUGGCUU ......(((((...........(((....)))..(((((......))))))))))((((..(((.((...............)))))..)))) ( -30.86) >DroEre_CAF1 27762 90 - 1 AAAUCAGCCGGAUCAUAAUUUAUUGGAGCCGAAAUGGG-GGAAUUUCCCACCGGCG-GCUGUGGACCACAAAAUGGACAAA-GGCCGUGGCUU .....((((((.((((..........(((((...((((-((.....))).))).))-))))))).)).....((((.....-..)))))))). ( -28.40) >DroYak_CAF1 32658 92 - 1 AAAUCAGCCGGAUCAUAAUUUAUUGGAGCCCAAAUGGGGGGAGUUUCCCACCGGCGAGCUGUGGGCCACAAAAUGAACAAA-GGCCGUGGCUU ......(((((.................(((....)))(((.....))).)))))((((..(((.((..............-)))))..)))) ( -32.34) >consensus AAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGGUGGAACUGCCCACCGGCGAGCUGUGGGCCACAAAAUGGACAAAAGGCCGUGGCUU ......(((((...........(((....)))..(((((......))))))))))(((((((((.((...............))))))))))) (-21.42 = -22.94 + 1.52)


| Location | 19,365,107 – 19,365,217 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -25.94 |
| Energy contribution | -28.67 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
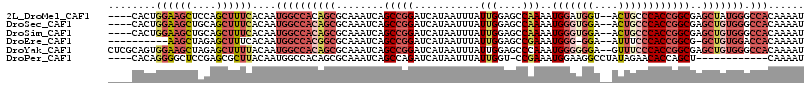
>2L_DroMel_CAF1 19365107 110 - 22407834 ----CACUGGAAGCUCCAGCUUUCACAAUGGCCACAGCGCAAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGAUGGU--ACUGCCCACCGGCGAGCUAUGGGCCACAAAAU ----...(((((((....)))))))...(((((.((..((...((.(((((......(((((((........)))))))((.--....))..)))))))))..)))))))...... ( -32.70) >DroSec_CAF1 28584 110 - 1 ----CACUGGAAGCUGCAGCUUUCACAAUGGCCACAGCGCAAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGGUGGA--ACUGCCCACCGGCGAGCUGUGGGCCACAAAAU ----...(((((((....)))))))...((((((((((........(((((...........(((....)))..(((((...--...))))))))))..))))).)))))...... ( -41.80) >DroSim_CAF1 28980 110 - 1 ----CACUGGAAGCUGCAGCUUUCACAAUGGCCACAGCGCAAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGGUGGA--ACUGCCCACCGGCGAGCUGUGGGCCACAAAAU ----...(((((((....)))))))...((((((((((........(((((...........(((....)))..(((((...--...))))))))))..))))).)))))...... ( -41.80) >DroEre_CAF1 27780 102 - 1 ----------AAGCUAGAGCUUUCACAAUGGCCACGGCGCAAAUCAGCCGGAUCAUAAUUUAUUGGAGCCGAAAUGGG-GGA--AUUUCCCACCGGCG-GCUGUGGACCACAAAAU ----------((((....))))......((((((((((........))).................(((((...((((-((.--....))).))).))-))))))).)))...... ( -35.30) >DroYak_CAF1 32676 114 - 1 CUCGCAGUGGAAGCUAGAGCUUUUACAAUGGCCACAGCGCAAAUCAGCCGGAUCAUAAUUUAUUGGAGCCCAAAUGGGGGGA--GUUUCCCACCGGCGAGCUGUGGGCCACAAAAU ......((((((((....))))))))..((((((((((........(((((.................(((....)))(((.--....))).)))))..))))).)))))...... ( -45.10) >DroPer_CAF1 47684 99 - 1 ----CACAGGGGCUCCGAGCGCUUACAAUGGCCACAGCGCAAAUCAGCCAGAUCAUAAUUUAUUGGU-CCGAAAUGGAAGGCCUAUAGAACACCAGCU------------CAAAAU ----....(((.(((((.(((((............)))))..........(((((........))))-).....))).)).)))..............------------...... ( -19.90) >consensus ____CACUGGAAGCUACAGCUUUCACAAUGGCCACAGCGCAAAUCAGCCGGAUCAUAAUUUAUUGGAGCCAAAAUGGGUGGA__ACUGCCCACCGGCGAGCUGUGGGCCACAAAAU ........((((((....))))))....((((((((((........(((((...........(((....)))..(((((((....))))))))))))..))))))).)))...... (-25.94 = -28.67 + 2.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:16 2006