
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,362,763 – 19,362,870 |
| Length | 107 |
| Max. P | 0.964908 |

| Location | 19,362,763 – 19,362,870 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.14 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -14.82 |
| Energy contribution | -13.38 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
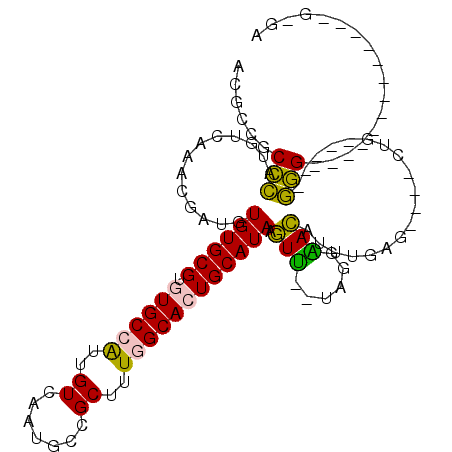
>2L_DroMel_CAF1 19362763 107 + 22407834 ACGACGCCCAUGUCAAACGAUGUGUGCGUGUGCCAUUGUCAAUGCCGCUUUGGCACUGCAUAAGUU--AAAGGACAUUUGUG----GUGUGGUGGGGGGGGGGGGGGGUUG-GU .((((.(((.......((((((.(((....)))))))))(..(.((.((.(.((((..((...(((--....)))...))..----)))).).)).)).)..)))).))))-.. ( -35.90) >DroPse_CAF1 39799 93 + 1 ACUUCUCUUUUGUCAAACGAUGUGUGCG--UGCCGUUGUCAAUGCCGCU-UAGCACUGCAUACAUAUGUAUGUAUGUCUGAAGAGACAA-----AAG-------------GACA ...((.((((((((.......(((.(((--(..........)))))))(-(((.(((((((((....))))))).))))))...)))))-----)))-------------)).. ( -26.90) >DroSec_CAF1 26262 89 + 1 ACACCGCCCAUGUCAAACGAUGUGUGCGUGUGCCAUUGUCAAUGCCGCUUUGGCACUGCAUAAGUU--UAGGAACAUUUGGG----CUG-----GGG-------------G-GG ...((.((((.(((((((....((((((.((((((..((.......))..)))))))))))).)))--).....(....)))----)))-----)).-------------)-). ( -32.90) >DroSim_CAF1 26625 89 + 1 ACGCCGCCCAUGUCAAACGAUGUGUGCGUGUGCCAUUGUCAAUGCCGCUUUGGCACUGCAUAAGUU--UAGGAACAUUUGGG----CUG-----GGG-------------G-GC ..(((.((((.(((((((....((((((.((((((..((.......))..)))))))))))).)))--).....(....)))----)))-----)).-------------)-)) ( -34.80) >DroPer_CAF1 44842 93 + 1 ACUUCUCUUUUGUCAAACGAUGUGUGCG--UGCCGUUGUCAAUGCUGCU-UAGCACUGCAUACAUAUGUAUGUAUGUCUGAAGAGACAA-----AAG-------------GACA ...((.((((((((.........((((.--.((.((.......)).)).-..)))).((((((((....)))))))).......)))))-----)))-------------)).. ( -26.80) >consensus ACGCCGCCCAUGUCAAACGAUGUGUGCGUGUGCCAUUGUCAAUGCCGCUUUGGCACUGCAUAAGUU__UAGGAACAUUUGAG____CUG_____GGG_____________G_GA ......(((.............((((((.((((((..((.......))..)))))))))))).(((......)))...................)))................. (-14.82 = -13.38 + -1.44)

| Location | 19,362,763 – 19,362,870 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.14 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -15.54 |
| Energy contribution | -14.66 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19362763 107 - 22407834 AC-CAACCCCCCCCCCCCCCCACCACAC----CACAAAUGUCCUUU--AACUUAUGCAGUGCCAAAGCGGCAUUGACAAUGGCACACGCACACAUCGUUUGACAUGGGCGUCGU ..-..................((.((.(----(....(((((....--(((...(((((((((.....))))))).)).((((....)).))....))).))))).)).)).)) ( -21.80) >DroPse_CAF1 39799 93 - 1 UGUC-------------CUU-----UUGUCUCUUCAGACAUACAUACAUAUGUAUGCAGUGCUA-AGCGGCAUUGACAACGGCA--CGCACACAUCGUUUGACAAAAGAGAAGU ..((-------------(((-----(((((........(((((((....)))))))(((((((.-...)))))))..(((((..--........))))).)))))))).))... ( -24.00) >DroSec_CAF1 26262 89 - 1 CC-C-------------CCC-----CAG----CCCAAAUGUUCCUA--AACUUAUGCAGUGCCAAAGCGGCAUUGACAAUGGCACACGCACACAUCGUUUGACAUGGGCGGUGU ..-(-------------(((-----((.----..((((((......--......(((((((((.....))))))).)).((((....)).))...))))))...)))).))... ( -25.90) >DroSim_CAF1 26625 89 - 1 GC-C-------------CCC-----CAG----CCCAAAUGUUCCUA--AACUUAUGCAGUGCCAAAGCGGCAUUGACAAUGGCACACGCACACAUCGUUUGACAUGGGCGGCGU ((-(-------------.((-----((.----..((((((......--......(((((((((.....))))))).)).((((....)).))...))))))...)))).))).. ( -30.10) >DroPer_CAF1 44842 93 - 1 UGUC-------------CUU-----UUGUCUCUUCAGACAUACAUACAUAUGUAUGCAGUGCUA-AGCAGCAUUGACAACGGCA--CGCACACAUCGUUUGACAAAAGAGAAGU ..((-------------(((-----(((((........(((((((....)))))))(((((((.-...)))))))..(((((..--........))))).)))))))).))... ( -24.00) >consensus UC_C_____________CCC_____CAG____CCCAAAUGUACAUA__AACUUAUGCAGUGCCAAAGCGGCAUUGACAAUGGCACACGCACACAUCGUUUGACAUGGGCGAAGU ..................................(((((...............(((((((((.....))))))).))...((....)).......)))))............. (-15.54 = -14.66 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:13 2006