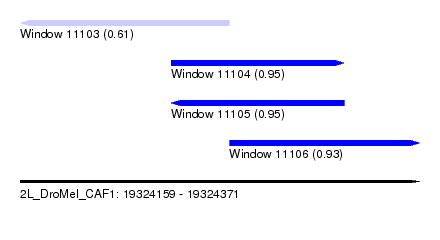
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,324,159 – 19,324,371 |
| Length | 212 |
| Max. P | 0.951693 |

| Location | 19,324,159 – 19,324,270 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -35.91 |
| Consensus MFE | -32.36 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19324159 111 - 22407834 ---------CCCAGCUCCCAGUGGCCAGCUUGGCCAUUUAGUGGCUGCCAGUCACGCUAUGAUUUGACCCACUUUUAUUGGUUCCAAAUAAGCCGACACGUAAUGAGCUCAAGGCUUAGC ---------..((((..(.(((((((.....)))))))..)..))))...(((..(((...(((((..(((.......)))...))))).))).)))......(((((.....))))).. ( -30.70) >DroSec_CAF1 16676 111 - 1 ---------CGCAGCUCCCAGUGGCCAGCUUGGCCAUUUAGUGGCUGCCAGUCACGCUAUGAUUUGACCCACUUUUAUUGGGUCCAAAUAAGCCGACACGUAAUGAGCUCAAGGCUUAGC ---------.(((((..(.(((((((.....)))))))..)..)))))..(((..(((...((((((((((.......))))).))))).))).)))......(((((.....))))).. ( -40.90) >DroSim_CAF1 18505 111 - 1 ---------CCCAGCUCCCAGUGGCCAGCUUGGCCAUUUAGUGGAUGCCAGUCACGCUAUGAUUUGACCCACUUUUAUUGGGUCCAAAUAAGCCGACACGUAAUGAGCUCAAGGCUUAGC ---------....(((((((((((((.....)))))))..).))).))..(((..(((...((((((((((.......))))).))))).))).)))......(((((.....))))).. ( -35.10) >DroEre_CAF1 15606 120 - 1 UCCUCGAUCCCCAGCUCCCAGUGGCCGGCUUGGCCAUUUAGUGGCUGCCAGUCACGCUAUGAUUUGACCCACUUUUAUUGGGACCGAAUAAGCCGACGCGUAAUGAGCUCAAGGCUUAGC ...(((.((((........(((((..(((..((((((...))))))))).((((..........)))))))))......)))).))).((((((...((.......))....)))))).. ( -36.94) >consensus _________CCCAGCUCCCAGUGGCCAGCUUGGCCAUUUAGUGGCUGCCAGUCACGCUAUGAUUUGACCCACUUUUAUUGGGUCCAAAUAAGCCGACACGUAAUGAGCUCAAGGCUUAGC ...........((((..(.(((((((.....)))))))..)..))))...(((..(((...((((((((((.......))))).))))).))).)))......(((((.....))))).. (-32.36 = -32.92 + 0.56)



| Location | 19,324,239 – 19,324,331 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.63 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19324239 92 + 22407834 UAAAUGGCCAAGCUGGCCACUGGGAGCUGGG----------------------------AAUCGGGAGUUAGCAGCUGAGAGAUGCUGGAUGCAUCGAGACCCCCAGUGGGGACAUAAAU .....((((.....))))(((((((((((..----------------------------((.(....)))..)))))....(((((.....)))))......))))))(....)...... ( -34.00) >DroSec_CAF1 16756 92 + 1 UAAAUGGCCAAGCUGGCCACUGGGAGCUGCG----------------------------AAUCGGGAGUUAGCGGCUGAGAGAUGCUGGAUGCAUUGAGACCCCCAUUGGGGACAUAAAU ....(((((.....))))).((((((((((.----------------------------...(....)...))))))....(((((.....)))))......)))).((....))..... ( -31.50) >DroSim_CAF1 18585 92 + 1 UAAAUGGCCAAGCUGGCCACUGGGAGCUGGG----------------------------AAUCGGGAGUUAGCAGCUGAGAGAUGCUGGAUGCAUCGAGACCCCCAGUGGGGACAUAAAU .....((((.....))))(((((((((((..----------------------------((.(....)))..)))))....(((((.....)))))......))))))(....)...... ( -34.00) >DroEre_CAF1 15686 120 + 1 UAAAUGGCCAAGCCGGCCACUGGGAGCUGGGGAUCGAGGAUCGGGAAUCGGCAACCGGGAACCGGGAGUUAGCAGCUAACCGAUGUUGGACGCAUCGAGACCCCCAGUGGGGCCAUAAAU ...((((((.......(((((((((((((((..(((.....)))...))(((..((((...))))..)))..)))))...((((((.....)))))).....)))))))))))))).... ( -47.11) >consensus UAAAUGGCCAAGCUGGCCACUGGGAGCUGGG____________________________AAUCGGGAGUUAGCAGCUGAGAGAUGCUGGAUGCAUCGAGACCCCCAGUGGGGACAUAAAU .....((((.....))))(((((((((((...........................................)))))....(((((.....)))))......))))))(....)...... (-27.94 = -27.63 + -0.31)

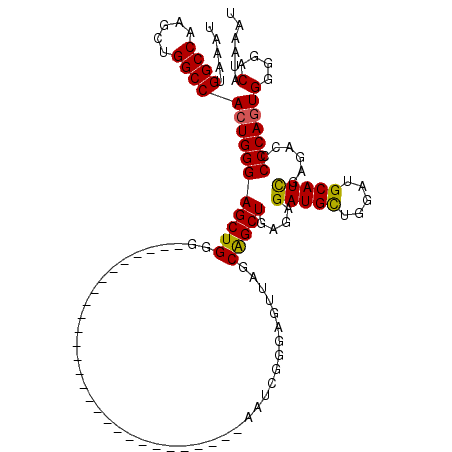
| Location | 19,324,239 – 19,324,331 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -22.59 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19324239 92 - 22407834 AUUUAUGUCCCCACUGGGGGUCUCGAUGCAUCCAGCAUCUCUCAGCUGCUAACUCCCGAUU----------------------------CCCAGCUCCCAGUGGCCAGCUUGGCCAUUUA .............(((((((((..((......((((........)))).....))..))))----------------------------))))).....(((((((.....))))))).. ( -28.30) >DroSec_CAF1 16756 92 - 1 AUUUAUGUCCCCAAUGGGGGUCUCAAUGCAUCCAGCAUCUCUCAGCCGCUAACUCCCGAUU----------------------------CGCAGCUCCCAGUGGCCAGCUUGGCCAUUUA ......(.((((....)))).)...((((.....)))).....(((.((............----------------------------.)).)))...(((((((.....))))))).. ( -23.22) >DroSim_CAF1 18585 92 - 1 AUUUAUGUCCCCACUGGGGGUCUCGAUGCAUCCAGCAUCUCUCAGCUGCUAACUCCCGAUU----------------------------CCCAGCUCCCAGUGGCCAGCUUGGCCAUUUA .............(((((((((..((......((((........)))).....))..))))----------------------------))))).....(((((((.....))))))).. ( -28.30) >DroEre_CAF1 15686 120 - 1 AUUUAUGGCCCCACUGGGGGUCUCGAUGCGUCCAACAUCGGUUAGCUGCUAACUCCCGGUUCCCGGUUGCCGAUUCCCGAUCCUCGAUCCCCAGCUCCCAGUGGCCGGCUUGGCCAUUUA ....(((((((((((((((((.((((.(.(((....((((((.(((((..(((.....)))..)))))))))))....))).)))))......))))))))))).......))))))... ( -45.21) >consensus AUUUAUGUCCCCACUGGGGGUCUCGAUGCAUCCAGCAUCUCUCAGCUGCUAACUCCCGAUU____________________________CCCAGCUCCCAGUGGCCAGCUUGGCCAUUUA ............(((((((((...(((((.....))))).......((........))...................................)))))))))((((.....))))..... (-22.59 = -23.15 + 0.56)


| Location | 19,324,270 – 19,324,371 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.77 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -31.31 |
| Energy contribution | -32.62 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19324270 101 + 22407834 -------------------AAUCGGGAGUUAGCAGCUGAGAGAUGCUGGAUGCAUCGAGACCCCCAGUGGGGACAUAAAUCCUUACGCUGCAGUUGAAAAACAAUUUUCCCCGGGUAUGC -------------------..(((((.....(((((((((.(((((.....)))))....((((....)))).........)))).))))).(((....))).......)))))...... ( -36.60) >DroSec_CAF1 16787 101 + 1 -------------------AAUCGGGAGUUAGCGGCUGAGAGAUGCUGGAUGCAUUGAGACCCCCAUUGGGGACAUAAAUCCUUUCGCUGCAGUUGAAAAACAAUUUUACCCGGGUAUGC -------------------..(((((.....(((((.(((((((((.....)))).....((((....)))).........)))))))))).(((....))).......)))))...... ( -34.10) >DroSim_CAF1 18616 101 + 1 -------------------AAUCGGGAGUUAGCAGCUGAGAGAUGCUGGAUGCAUCGAGACCCCCAGUGGGGACAUAAAUCCUUACGCUGCAGUUGAAAAACAAUUUUGCCCGGGUAUGC -------------------..(((((.....(((((((((.(((((.....)))))....((((....)))).........)))).))))).(((....))).......)))))...... ( -36.50) >DroEre_CAF1 15726 120 + 1 UCGGGAAUCGGCAACCGGGAACCGGGAGUUAGCAGCUAACCGAUGUUGGACGCAUCGAGACCCCCAGUGGGGCCAUAAAUCCUUACGCUGCAGUUGGAAAACAAUUUUCGCCGGGUAUGC ..........((((((.((...((..((((.(((((....((((((.....)))))).(.((((....)))).)............))))).(((....)))))))..)))).))).))) ( -39.40) >consensus ___________________AAUCGGGAGUUAGCAGCUGAGAGAUGCUGGAUGCAUCGAGACCCCCAGUGGGGACAUAAAUCCUUACGCUGCAGUUGAAAAACAAUUUUCCCCGGGUAUGC .....................(((((.....(((((((((.(((((.....)))))....((((....)))).........)))).))))).(((....))).......)))))...... (-31.31 = -32.62 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:03 2006