
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,323,239 – 19,323,387 |
| Length | 148 |
| Max. P | 0.990772 |

| Location | 19,323,239 – 19,323,357 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -31.94 |
| Energy contribution | -31.88 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19323239 118 + 22407834 AAACUUUUGGUGCAACUCUGCGGGAAAAACGGUUGAAUAA-AAUGCAGGCA-UUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACC ....((((..((((....))))..))))...((((.....-...((.(((.-.((.....))))).))((((((((((((((.((..((....))..))))))))))))))))))))... ( -36.70) >DroSec_CAF1 15733 120 + 1 AAACUUUUGGUCCAACUCUGCGGAAAAAACGGUUGAAUAAAAAUGCAGGGAUUUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACC ...(.(((..(((........)))..))).)((((.........((...((((.(.....))))).))((((((((((((((.((..((....))..))))))))))))))))))))... ( -35.50) >DroSim_CAF1 17561 119 + 1 AAACUUUUGGUGCAACUCUGCGGGAAAAACGGUUGCAUAAAAAUGCAGGAA-UUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACC ........((((.......((.((....((((((((((....)))))...)-)))).......)).))((((((((((((((.((..((....))..))))))))))))))))...)))) ( -39.20) >DroEre_CAF1 14737 117 + 1 AAACU-UUGGUGCAGCUCUGCAGGAAAAGCGGUUGAAUAAAAAUGCAGAAA-UUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCG-GGCACAACAACUGCAAUUGGCCAACACC .....-..((((..((((((((.....................)))))).(-(((.....))))..))((((((((((((((.((..((..-.))..))))))))))))).)))..)))) ( -32.20) >consensus AAACUUUUGGUGCAACUCUGCGGGAAAAACGGUUGAAUAAAAAUGCAGGAA_UUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACC ........(((((((.((((((.....................))))))...)))...........(.((((((((((((((.((..((....))..)))))))))))))))))..)))) (-31.94 = -31.88 + -0.06)

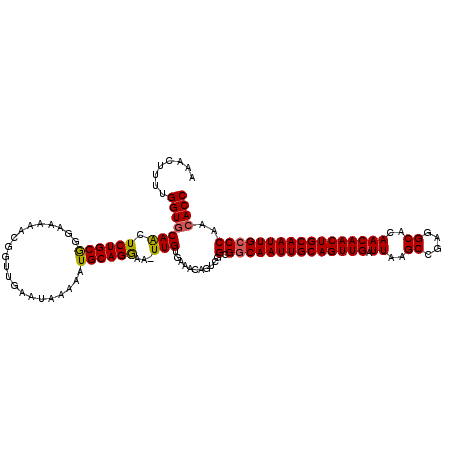
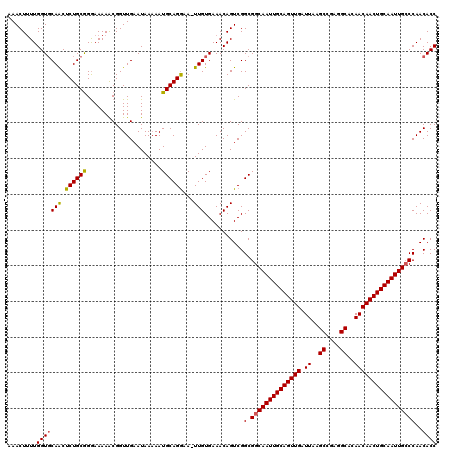
| Location | 19,323,239 – 19,323,357 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -35.00 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19323239 118 - 22407834 GGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAA-UGCCUGCAUU-UUAUUCAACCGUUUUUCCCGCAGAGUUGCACCAAAAGUUU ((((...(((((((((((((((((.((....)).))).)))))))))))))).............(((-(..((((...-...................)))).))))))))........ ( -39.35) >DroSec_CAF1 15733 120 - 1 GGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAAAUCCCUGCAUUUUUAUUCAACCGUUUUUUCCGCAGAGUUGGACCAAAAGUUU (((....(((((((((((((((((.((....)).))).)))))))))))))).(((((((.....)......((((.......................)))))))))))))........ ( -40.10) >DroSim_CAF1 17561 119 - 1 GGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAA-UUCCUGCAUUUUUAUGCAACCGUUUUUCCCGCAGAGUUGCACCAAAAGUUU (((((..(((((((((((((((((.((....)).))).)))))))))))))).....((((.......-....(((((....)))))............))))....)))))........ ( -42.10) >DroEre_CAF1 14737 117 - 1 GGUGUUGGCCAAUUGCAGUUGUUGUGCC-CGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAA-UUUCUGCAUUUUUAUUCAACCGCUUUUCCUGCAGAGCUGCACCAA-AGUUU ((((((((((((((((((((((((.((.-..)).))).))))))))))))..))))))..........-.(((((((.....................)))))))....)))..-..... ( -35.80) >consensus GGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAA_UUCCUGCAUUUUUAUUCAACCGUUUUUCCCGCAGAGUUGCACCAAAAGUUU (((((..(((((((((((((((((.((....)).))).))))))))))))))....................((((.......................))))....)))))........ (-35.00 = -35.50 + 0.50)

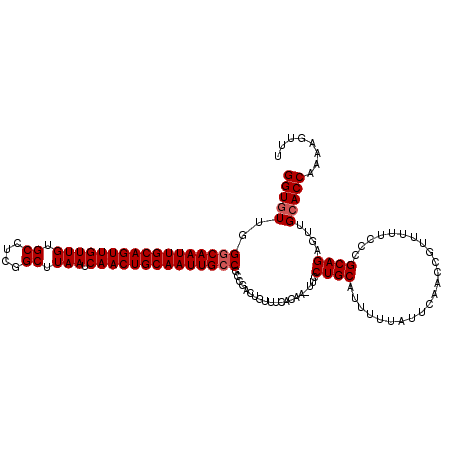
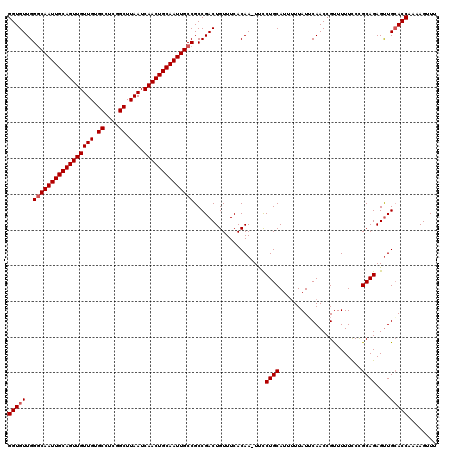
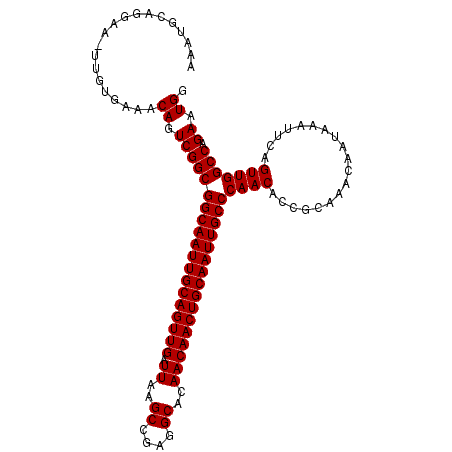
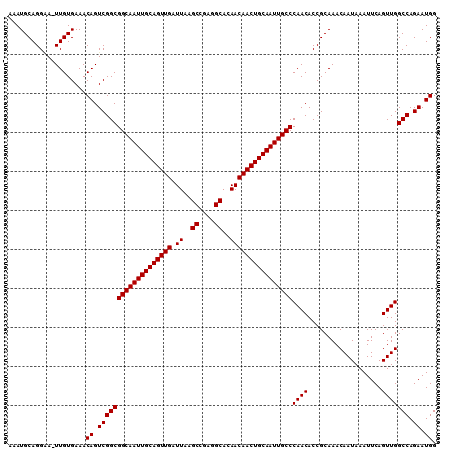
| Location | 19,323,279 – 19,323,387 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 97.87 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -33.81 |
| Energy contribution | -33.81 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19323279 108 + 22407834 -AAUGCAGGCA-UUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACCGCAAACAAUAAAUUCAGUUGGCCAGAAUGG -......(((.-(((((.....(....)((((((((((((((.((..((....))..))))))))))))))))......))))).((((.......)))))))....... ( -35.00) >DroSec_CAF1 15773 110 + 1 AAAUGCAGGGAUUUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACCGCAAACAAUAAAUUCAGUUGGCCAGAAUGG ....((..((((((((......(.(((.((((((((((((((.((..((....))..)))))))))))))))).....)))).....)))))))).....))........ ( -34.00) >DroSim_CAF1 17601 109 + 1 AAAUGCAGGAA-UUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACCGCAAACAAUAAAUUCAGUUGGCCAGAAUGG ...........-........((.(((((((((((((((((((.((..((....))..))))))))))))))))((((...................))))))).)).)). ( -33.81) >consensus AAAUGCAGGAA_UUGUGAAACAGUCGGCGGCAAUUGCAGUUGAUUAAGCCGAGGCACAACAACUGCAAUUGCCCAACACCGCAAACAAUAAAUUCAGUUGGCCAGAAUGG ....................((.(((((((((((((((((((.((..((....))..))))))))))))))))((((...................))))))).)).)). (-33.81 = -33.81 + -0.00)

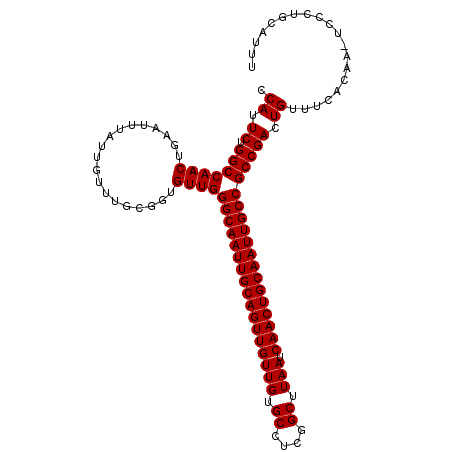
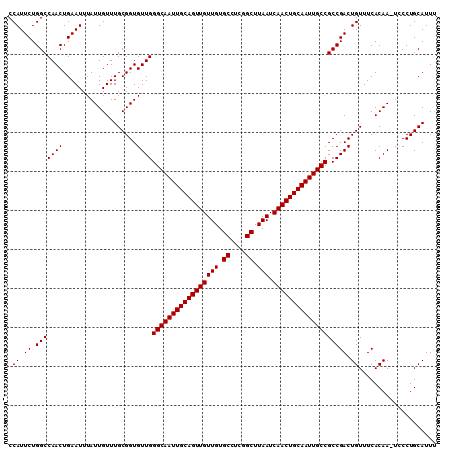
| Location | 19,323,279 – 19,323,387 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 97.87 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -35.31 |
| Energy contribution | -35.31 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19323279 108 - 22407834 CCAUUCUGGCCAACUGAAUUUAUUGUUUGCGGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAA-UGCCUGCAUU- ..((((.(.....).))))........(((((((((((((((((((((((((((.((....)).))).))))))))))))))...((.....)).)))-)))).)))..- ( -39.40) >DroSec_CAF1 15773 110 - 1 CCAUUCUGGCCAACUGAAUUUAUUGUUUGCGGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAAAUCCCUGCAUUU .((.((.(((((((...................))))(((((((((((((((((.((....)).))).))))))))))))))))))).)).................... ( -35.31) >DroSim_CAF1 17601 109 - 1 CCAUUCUGGCCAACUGAAUUUAUUGUUUGCGGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAA-UUCCUGCAUUU ..((((.(.....).))))........(((((.(((((((((((((((((((((.((....)).))).))))))))))))))...((.....)).)))-)..)))))... ( -36.30) >consensus CCAUUCUGGCCAACUGAAUUUAUUGUUUGCGGUGUUGGGCAAUUGCAGUUGUUGUGCCUCGGCUUAAUCAACUGCAAUUGCCGCCGACUGUUUCACAA_UCCCUGCAUUU .((.((.(((((((...................))))(((((((((((((((((.((....)).))).))))))))))))))))))).)).................... (-35.31 = -35.31 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:59 2006