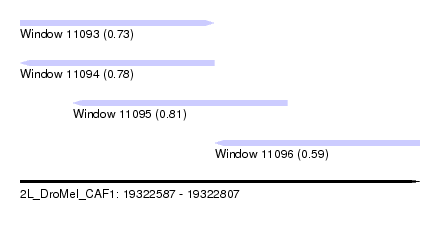
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,322,587 – 19,322,807 |
| Length | 220 |
| Max. P | 0.808015 |

| Location | 19,322,587 – 19,322,694 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19322587 107 + 22407834 ---AUAAGUAUUU-CAGUGUAAAACUGCGAAUGCAACUUUCUGCAUUUUGAUGCGGCAGUUAUUCGACUUUAUCGUGUGUGCUGACACGAAAUAUUUUAGCAUUCUAUUUA ---.((((((...-((((.....)))).((((((((((..((((((....)))))).))))...........((((((......)))))).........)))))))))))) ( -27.20) >DroSec_CAF1 15075 111 + 1 AUGAUAAUUAUUUCCAGUGUAAAGCUGCGAAUGCAACUUUUUGCAUUUUGAUGCGGCAGUUAUUCGACUUUAUCGUGUGUGCUGACACGAAAUAUUUUAGCAUUCUAUUUA (((..((.((((((..((((...(((((((((((((....))))))))....)))))((((((.(((.....))).))).))).)))))))))).))...)))........ ( -24.50) >DroSim_CAF1 16903 111 + 1 AUGAUAAUUAUUUCCAGUGGAAAACUGCGAAUGCAACUUUCUGCAUUUUGAUGCGGCAGUUAUUCGACUUUAUCUUGUGUGCUGACACGAAAUAUUUUAGCAUUCUAUUUA (((..((.((((((.(((.((((((((((((((((......))))))).......)))))).))).))).......((((....)))))))))).))...)))........ ( -24.91) >DroEre_CAF1 14104 111 + 1 AUUUUCAGUGUUUUCAGUGUAAGCCUGCGAAUGCAAGUUUCUGCAUUUUUAUGCGGCAGUUAUUCGACUUUAUCGUGUGUGCUGACACGAAGUAUUUUAGCAUUCUAUUUA .....(((.((((.......))))))).((((((......((((((....))))))..........(((...((((((......)))))))))......))))))...... ( -24.90) >consensus AUGAUAAGUAUUUCCAGUGUAAAACUGCGAAUGCAACUUUCUGCAUUUUGAUGCGGCAGUUAUUCGACUUUAUCGUGUGUGCUGACACGAAAUAUUUUAGCAUUCUAUUUA ..............((((.....)))).((((((((((..((((((....)))))).))))...........((((((......)))))).........))))))...... (-22.24 = -22.68 + 0.44)

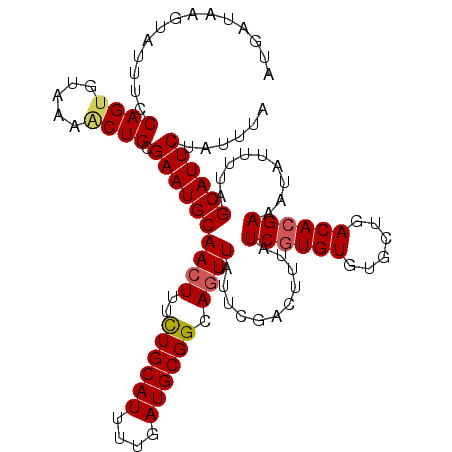
| Location | 19,322,587 – 19,322,694 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19322587 107 - 22407834 UAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAGAAAGUUGCAUUCGCAGUUUUACACUG-AAAUACUUAU--- ......((((((.........((((((......))))))...........((((..(((((....)))).)..)))))))))).((((.....))))-..........--- ( -21.50) >DroSec_CAF1 15075 111 - 1 UAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAAAAAGUUGCAUUCGCAGCUUUACACUGGAAAUAAUUAUCAU ......((.............((((((......))))))................((((((....))))..((((((((....)))))))).....)).........)).. ( -20.40) >DroSim_CAF1 16903 111 - 1 UAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACAAGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAGAAAGUUGCAUUCGCAGUUUUCCACUGGAAAUAAUUAUCAU .................((((((((((....)))).......(((.(((.((((((.((((....)))).(((.......)))))))))))).))).))))))........ ( -21.40) >DroEre_CAF1 14104 111 - 1 UAAAUAGAAUGCUAAAAUACUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUAAAAAUGCAGAAACUUGCAUUCGCAGGCUUACACUGAAAACACUGAAAAU ....................(((((((.(((.....((((...)))).....((((.((((....)))).(((.......)))))))))).)))).)))............ ( -20.30) >consensus UAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAGAAAGUUGCAUUCGCAGUUUUACACUGGAAAUAAUUAUCAU ......((((((.........((((((......))))))..................((((....))))........)))))).(((.......))).............. (-17.83 = -18.07 + 0.25)

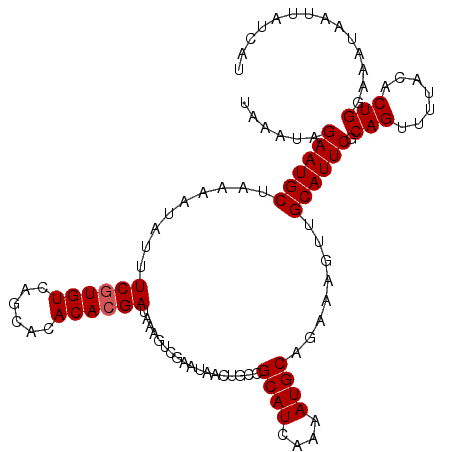
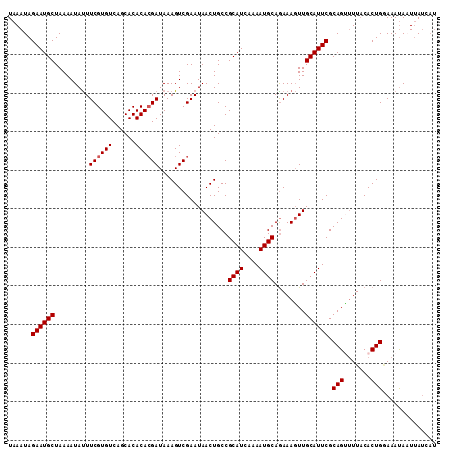
| Location | 19,322,616 – 19,322,734 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.97 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19322616 118 - 22407834 UUUCGGGGAAACCCACCGUGGAAUCGCUUUAAAUGUGCAUUAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAGAAAGUUG (((((((....)))...((((..((((((((.....(((((......))))).........((((((......))))))))))).))).......))))...........)))).... ( -30.70) >DroSec_CAF1 15108 118 - 1 UUUCGGGGAAACCCCCCGUGGAACCGCUUUAAAUGUGCAUUAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAAAAAGUUG ...(((((.....)))))......(((((((.....(((((......))))).........((((((......))))))))))).))..(((((...((((....))))....))))) ( -28.60) >DroSim_CAF1 16936 118 - 1 UUUCGGGGAAACCCACCGUGGAAUCGCUUUAAAUGUGCAUUAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACAAGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAGAAAGUUG (((((((....)))...((((..((((((((..((((((((......)))))...........(((....))))))...))))).))).......))))...........)))).... ( -27.80) >DroEre_CAF1 14137 117 - 1 UUUCGGGGAAACC-AUCACCGAGUCGCUUUAAACGUGCAUUAAAUAGAAUGCUAAAAUACUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUAAAAAUGCAGAAACUUG .....((....))-.....((((((((((((.....(((((......))))).........((((((......))))))))))).))).........((((....)))).....)))) ( -26.20) >consensus UUUCGGGGAAACCCACCGUGGAAUCGCUUUAAAUGUGCAUUAAAUAGAAUGCUAAAAUAUUUCGUGUCAGCACACACGAUAAAGUCGAAUAACUGCCGCAUCAAAAUGCAGAAAGUUG ....(((....))).........((((((((.....(((((......))))).........((((((......))))))))))).))).(((((...((((....))))....))))) (-23.98 = -24.97 + 1.00)

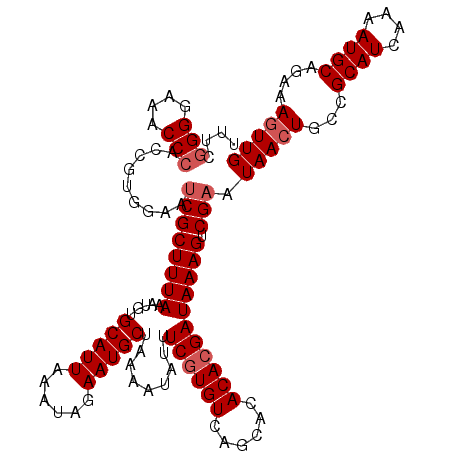
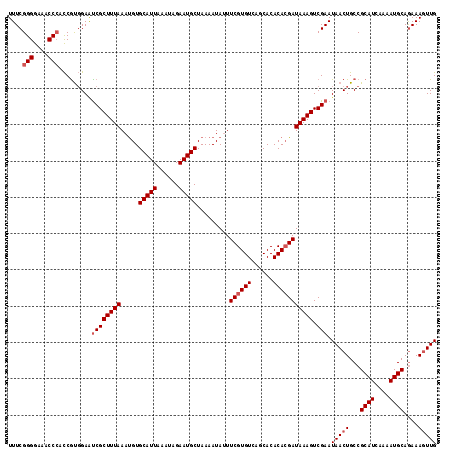
| Location | 19,322,694 – 19,322,807 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.28 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -29.21 |
| Energy contribution | -29.09 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19322694 113 - 22407834 ACACUCGGCUUUGCGGUUAGCAGCUGGAGGCUGCCAUCUCCUCCAGGAAUGCAUCUUGGGAUCACGCACAGU-------CUUUCGGGGAAACCCACCGUGGAAUCGCUUUAAAUGUGCAU .(((..((((.((((....(((((.....))))).......(((((((.....)))))))....)))).)))-------)....(((....)))...))).....((.........)).. ( -38.70) >DroSec_CAF1 15186 120 - 1 ACACUCGGCUUUGCGGUUAGCAGCUGGAGGCUGCCAUCUCCUCCAGGAGUGCAUCUUGGGAUCACGCACAGUCGCACAUCUUUCGGGGAAACCCCCCGUGGAACCGCUUUAAAUGUGCAU ......((((.((((....(((((.....))))).......(((((((.....)))))))....)))).))))((((((.....((((.....))))(((....))).....)))))).. ( -46.30) >DroSim_CAF1 17014 120 - 1 ACACUCGGCUUUGCGGUUAGCAGCUGGAGGCUGCCAUCUCCUCCAGGAGUGCAUCUCGGGAUCACGCACAGUCGCACAUCUUUCGGGGAAACCCACCGUGGAAUCGCUUUAAAUGUGCAU ......((((.((((........((.((((.(((...((((....)))).))))))).))....)))).))))((((((.....(((....)))...(((....))).....)))))).. ( -43.50) >DroEre_CAF1 14215 108 - 1 GCACUCGACUUUGCGGUUAGCAGCUGGAGGCUGUCUGCUCCUCCAGGAAUGCAUCUC-----------CAGUCGAACAUCUUUCGGGGAAACC-AUCACCGAGUCGCUUUAAACGUGCAU ((((((((((....((...(((.(((((((..(....).)))))))...)))....)-----------)))))))......(((((.((....-.)).)))))...........)))).. ( -34.70) >consensus ACACUCGGCUUUGCGGUUAGCAGCUGGAGGCUGCCAUCUCCUCCAGGAAUGCAUCUCGGGAUCACGCACAGUCGCACAUCUUUCGGGGAAACCCACCGUGGAAUCGCUUUAAAUGUGCAU .(((........((((((.(((.(((((((.........)))))))...)))................................(((....))).......)))))).......)))... (-29.21 = -29.09 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:55 2006