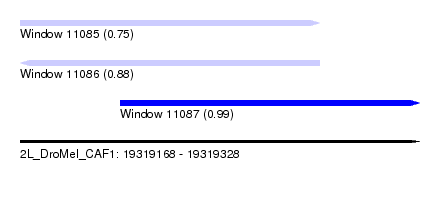
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,319,168 – 19,319,328 |
| Length | 160 |
| Max. P | 0.989836 |

| Location | 19,319,168 – 19,319,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19319168 120 + 22407834 GUAAGGCGAUUAUAAAAUUAAUUCGAGCGGGCUAUAAAACACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAA .....(((((((......)))))...))(((((((.................((....(((((.((((.....)))).)).)))..))....(((((((....))))))).))))))).. ( -29.30) >DroSec_CAF1 11759 120 + 1 GUGAGGCGAUUAUAAAAUUAAUUCGAGCGGGCUAUAAAACACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAA .(((((((((((......)))))..((.(((((...............((((.......)))).((((.....)))).))))).))))))))..(((((((((......))))..))))) ( -33.00) >DroSim_CAF1 13585 120 + 1 GUGAGGCGAUUAUAAAAUUAAUUCGAGCGGGCUAUAAAACACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAA .(((((((((((......)))))..((.(((((...............((((.......)))).((((.....)))).))))).))))))))..(((((((((......))))..))))) ( -33.00) >DroEre_CAF1 10896 119 + 1 GUGAGGCGAUUAUGAAUUUAAUGCGAGCAGACUAUAAAACACUUAAUAAGCUGCUUCCUGGCCAAGUGAAAAUCACUGGGUGCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGC-CAA .(((((((((((......))))((((((((..(((.((....)).)))..))))))....(((.((((.....)))).)))))..)))))))(((((((....)))))))......-... ( -33.40) >DroYak_CAF1 12029 120 + 1 GUGAGGCGAUUAUGAAAUUAAUUCGAGCGGACUAUAAAACACUUAAUAAGGUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACGGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAA .(((((((((((......)))))...(.(((((......((((((...(((.....)))...))))))..........))))).).))))))..(((((((((......))))..))))) ( -33.49) >consensus GUGAGGCGAUUAUAAAAUUAAUUCGAGCGGGCUAUAAAACACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAA .((((((.................((((((..(((.((....)).)))..))))))...((((.((((.....)))).))))....))))))(((((((....))))))).......... (-27.88 = -28.16 + 0.28)

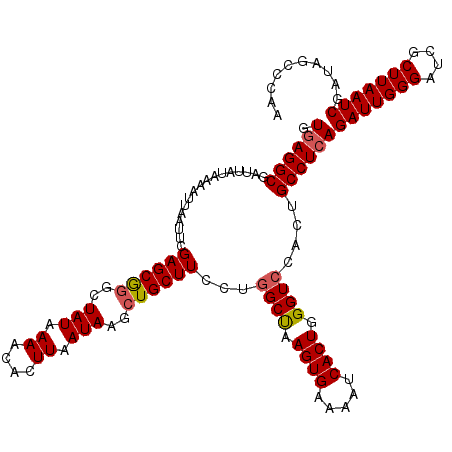
| Location | 19,319,168 – 19,319,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.880945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19319168 120 - 22407834 UUGGGCUAUCGAUUAAGCGAUCCCAAUCUGAGGCAGUGGACCCAGUGAUUUUCACUUAGCCAGGAAGCAGCUUAUUAAGUGUUUUAUAGCCCGCUCGAAUUAAUUUUAUAAUCGCCUUAC (((((..((((......)))))))))..(((((((((((....((((.....))))..((...((((((.((.....))))))))...))))))).((.(((......))))))))))). ( -29.80) >DroSec_CAF1 11759 120 - 1 UUGGGCUAUCGAUUAAGCGAUCCCAAUCUGAGGCAGUGGACCCAGUGAUUUUCACUUAGCCAGGAAGCAGCUUAUUAAGUGUUUUAUAGCCCGCUCGAAUUAAUUUUAUAAUCGCCUCAC (((((..((((......)))))))))..(((((((((((....((((.....))))..((...((((((.((.....))))))))...))))))).((.(((......))))))))))). ( -32.00) >DroSim_CAF1 13585 120 - 1 UUGGGCUAUCGAUUAAGCGAUCCCAAUCUGAGGCAGUGGACCCAGUGAUUUUCACUUAGCCAGGAAGCAGCUUAUUAAGUGUUUUAUAGCCCGCUCGAAUUAAUUUUAUAAUCGCCUCAC (((((..((((......)))))))))..(((((((((((....((((.....))))..((...((((((.((.....))))))))...))))))).((.(((......))))))))))). ( -32.00) >DroEre_CAF1 10896 119 - 1 UUG-GCUAUCGAUUAAGCGAUCCCAAUCUGAGGCAGUGCACCCAGUGAUUUUCACUUGGCCAGGAAGCAGCUUAUUAAGUGUUUUAUAGUCUGCUCGCAUUAAAUUCAUAAUCGCCUCAC (((-(..((((......)))).))))..(((((((((((.((.((((.....)))).))....((.((((.((((..........)))).)))))))))))............)))))). ( -30.10) >DroYak_CAF1 12029 120 - 1 UUGGGCUAUCGAUUAAGCGAUCCCAAUCUGAGGCCGUGGACCCAGUGAUUUUCACUUAGCCAGGAAGCACCUUAUUAAGUGUUUUAUAGUCCGCUCGAAUUAAUUUCAUAAUCGCCUCAC (((((..((((......)))))))))..((((((.((((((...((((....(((((((..(((.....)))..)))))))..)))).))))))..(((.....)))......)))))). ( -37.80) >consensus UUGGGCUAUCGAUUAAGCGAUCCCAAUCUGAGGCAGUGGACCCAGUGAUUUUCACUUAGCCAGGAAGCAGCUUAUUAAGUGUUUUAUAGCCCGCUCGAAUUAAUUUUAUAAUCGCCUCAC (((((..((((......)))))))))..((((((.((((....((((.....))))..((...((((((.((.....))))))))...))))))..(.((((......))))))))))). (-27.44 = -27.28 + -0.16)

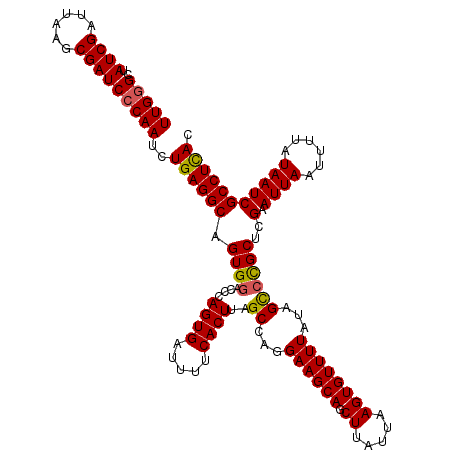
| Location | 19,319,208 – 19,319,328 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19319208 120 + 22407834 ACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAAGUACAAGCUCGUUAAGAGCACACGAAGCAAACAAAGAUCA ...........((((((..(((((((((.....))))((((.....))))..(((((((....)))))))..))))).........((((.....))))....))))))........... ( -31.80) >DroSec_CAF1 11799 120 + 1 ACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAAGUACAAGCUCGUUAAGAGCACACGAAGCAAACAAAGAUCA ...........((((((..(((((((((.....))))((((.....))))..(((((((....)))))))..))))).........((((.....))))....))))))........... ( -31.80) >DroSim_CAF1 13625 120 + 1 ACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAAGUACAAGCUCGUUAAGAGCACACGAAGCAAACAAAGAUCA ...........((((((..(((((((((.....))))((((.....))))..(((((((....)))))))..))))).........((((.....))))....))))))........... ( -31.80) >DroEre_CAF1 10936 119 + 1 ACUUAAUAAGCUGCUUCCUGGCCAAGUGAAAAUCACUGGGUGCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGC-CAAGUACAAGCUCGUUAAGAGCACACGAGGCAAACAAAGAUCA .(((....((.(((......(((.((((.....)))).))))))))(((((...((((.((((......))))..)-)))......((((.....))))....))))).....))).... ( -32.70) >DroYak_CAF1 12069 120 + 1 ACUUAAUAAGGUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACGGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAAGUACAAGCUCGUUAAGAGCACACGAAGCAAACAAAGAUCA ...........((((((..(((((((((.....))))((((.....))))..(((((((....)))))))..))))).........((((.....))))....))))))........... ( -33.60) >consensus ACUUAAUAAGCUGCUUCCUGGCUAAGUGAAAAUCACUGGGUCCACUGCCUCAGAUUGGGAUCGCUUAAUCGAUAGCCCAAGUACAAGCUCGUUAAGAGCACACGAAGCAAACAAAGAUCA ...........((((((..(((((((((.....))))((((.....))))..(((((((....)))))))..))))).........((((.....))))....))))))........... (-30.40 = -30.64 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:46 2006