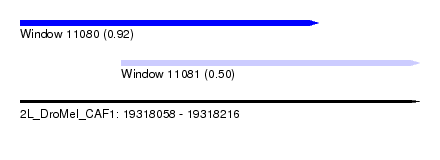
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,318,058 – 19,318,216 |
| Length | 158 |
| Max. P | 0.916728 |

| Location | 19,318,058 – 19,318,176 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19318058 118 + 22407834 CUCUGCAGGUAGCCAAAAUCCCGAUCCCCAACUCAAAAUACAAAAUACAAAAUACAGGCCCA--AUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCG .......((.((((........((........)).......................(((((--((.(....)(((.....)))))))))))))))).((..((((.....))))..)). ( -24.30) >DroSec_CAF1 10616 120 + 1 CUCCGCAGGUAGCCAAAAUCCCGAUCCCCAACUCAAAAUACAAAAUACAAAAUACAGGCCCAAAAUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCG .......((.((((........((........)).......................((((((....(....)(((.....))).)))))))))))).((..((((.....))))..)). ( -23.90) >DroSim_CAF1 12473 120 + 1 CUCCGCAGGUAGCCAAAAUCCCGAUCCCCAAAUCAAAAUACAAAAUACAAAAUACAGGCCCAAAAUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCG .......((.((((........(((......))).......................((((((....(....)(((.....))).)))))))))))).((..((((.....))))..)). ( -25.10) >DroEre_CAF1 9841 117 + 1 CUCUGCAGGUAGCCAAAAUCCCGAUCCCCAACUCAAAAUACAAAAUACAAAAUACAGGCCCA--AUCG-GCCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCG .......((.((((.....((((((...............................((((..--...)-))).(((.....))))))))).)))))).((..((((.....))))..)). ( -26.80) >DroYak_CAF1 10886 115 + 1 CUCUGCAGGUAGCCAAAAUC-CGAUCCCCAACUCAAAAUACA--AUCCCAAAUACAGGCCCU--AUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCG .......((.((((......-.((........))........--..(((((.....(((.(.--...).))).(((.....))).))))).)))))).((..((((.....))))..)). ( -21.90) >consensus CUCUGCAGGUAGCCAAAAUCCCGAUCCCCAACUCAAAAUACAAAAUACAAAAUACAGGCCCA__AUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCG .......((.((((........((........)).......................((((......(....)(((.....)))...)))))))))).((..((((.....))))..)). (-21.60 = -21.60 + -0.00)

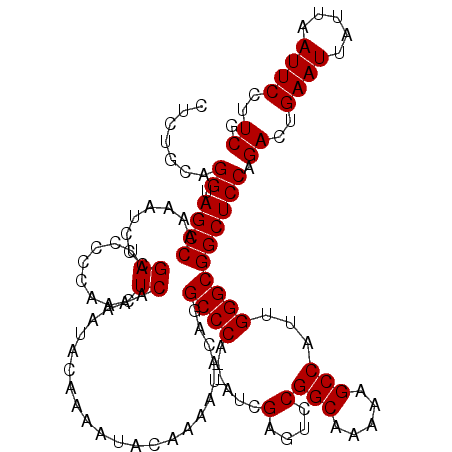
| Location | 19,318,098 – 19,318,216 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19318098 118 + 22407834 CAAAAUACAAAAUACAGGCCCA--AUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCGGCCCGCUUGGCCGUCUGCACUGCAGGUGAUUUUGCUAACA ((((((.((.....((((((((--((.(....)(((.....)))))))))).....(((((.((((.....))))....((((.....)))))))))..)))....))))))))...... ( -33.00) >DroSec_CAF1 10656 120 + 1 CAAAAUACAAAAUACAGGCCCAAAAUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCGGCCCGCUUGGCCGUCUGCACUGCAGGUGAUUUUGCUAACA ((((((.((.....(((((((((....(....)(((.....))).)))))).....(((((.((((.....))))....((((.....)))))))))..)))....))))))))...... ( -32.60) >DroSim_CAF1 12513 120 + 1 CAAAAUACAAAAUACAGGCCCAAAAUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCGGCCCGCUUGGCCGUCUGCACUGCAGGUGAUUUUGCUAACA ((((((.((.....(((((((((....(....)(((.....))).)))))).....(((((.((((.....))))....((((.....)))))))))..)))....))))))))...... ( -32.60) >DroEre_CAF1 9881 117 + 1 CAAAAUACAAAAUACAGGCCCA--AUCG-GCCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCGGCCCGCUUGGCCGUCUGCACUGCAGGUGAUUUUGCUAACA ................((((..--...)-))).(((((((.((((.((((((....).((..((((.....))))..)).)))))..))))(..((((...))))..).))))))).... ( -35.20) >DroYak_CAF1 10925 115 + 1 CA--AUCCCAAAUACAGGCCCU--AUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCGGC-CGUUUGGCCGUCUGCACUGCAGGUGAUUUUGCUAACA .(--(((((........((((.--...(....)(((.....)))...)))).((..(((((.((((.....))))....(((-(....)))))))))....)).)).))))......... ( -31.10) >consensus CAAAAUACAAAAUACAGGCCCA__AUCGAGUCCGGCAAAAAGCCAUUGGGCGGCUCCAGACUGAAUUAUUAAUUCCUUCGGCCCGCUUGGCCGUCUGCACUGCAGGUGAUUUUGCUAACA ................(((.(......).))).(((((((.(((.....((((...(((((.((((.....))))....((((.....)))))))))..)))).)))..))))))).... (-28.90 = -28.94 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:40 2006