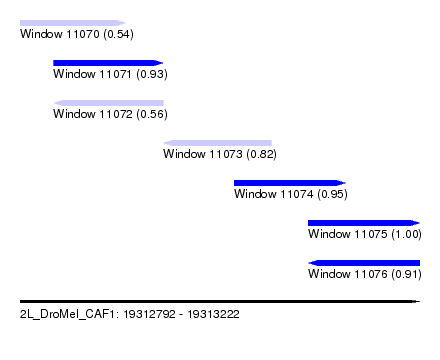
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,312,792 – 19,313,222 |
| Length | 430 |
| Max. P | 0.997601 |

| Location | 19,312,792 – 19,312,906 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19312792 114 + 22407834 AGCAAGGGGGUGCAACUAAAAACUGAAACUUAUAC----UA--ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUUGGGUCACAAAAGACAACACAGAAAUUCCUUAGCGG .((.(((((((....((((((..............----..--.(((((.....))))).........((((.....))))))))))(((......))).........))))))).)).. ( -27.20) >DroSec_CAF1 5673 114 + 1 AGCAAGGGGCUGCAACUAAAAACCGAAACUUAUAC----UA--ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGAGCACAAAAGACAACACAGAAAUUCCUUAGCGG .(((((((.(((..........((((((.......----..--.(((((.....))))).........((((.....)))))))))).((......).)....)))....))))).)).. ( -25.50) >DroSim_CAF1 6156 114 + 1 AGCAAGGGGCUGCAACUAAAAACCGAAACUUAUAC----UA--ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGGGCACAAAAGACAAAACAGAAAUUCCUUAGUGG .(((((((.(((..........((((((.......----..--.(((((.....))))).........((((.....))))))))))(..........)....)))....))))).)).. ( -23.40) >DroEre_CAF1 5067 117 + 1 AGCAGGGG-GUGCGACUAAAAACUGAAACUUAUGCAUACUUAUACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGGGC--AAAAGACAAUACACAAAUUCCUUACCAG ....((((-(((((..................))))).......(((((.....)))))...))))..((((((((((((((....))).--))))))..........)))))....... ( -25.37) >DroYak_CAF1 5669 119 + 1 GGCAAGGG-GUGCAACUAAAAACUGAAACUUAUACAUACUAAUACCCCCACAAUUGGGGAAAUCCUACGGAAUUCUUUUCCUUUCGGGGCACAAAAGACAAUACCAAAAUUCCUUACCAG ((.(((((-((((.........((((((.................(((((....))))).........((((.....)))))))))).))))....(.......).....))))).)).. ( -26.00) >consensus AGCAAGGGGGUGCAACUAAAAACUGAAACUUAUAC____UA__ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGGGCACAAAAGACAACACAGAAAUUCCUUAGCGG .((.(((((((...........((((((................(((((.....))))).........((((.....)))))))))).........(......)....))))))).)).. (-19.26 = -19.94 + 0.68)

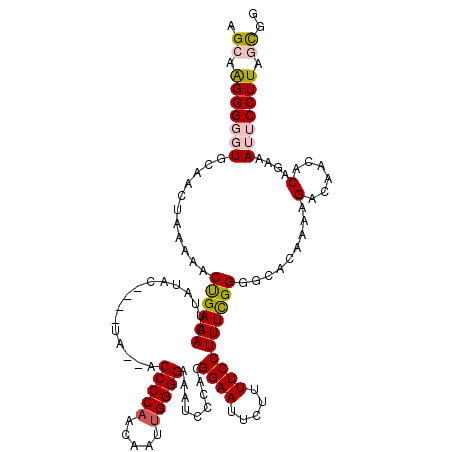
| Location | 19,312,828 – 19,312,946 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.29 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19312828 118 + 22407834 A--ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUUGGGUCACAAAAGACAACACAGAAAUUCCUUAGCGGACGCAGUGAAAAAUAUUAAAUUUUACCUCCACACUGGGGC .--.(((((.....)))))...(((((.((((((((....((((((.....)))))).......)).))))))...(.(((....((((((.........)))))).))).)..))))). ( -28.20) >DroSec_CAF1 5709 118 + 1 A--ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGAGCACAAAAGACAACACAGAAAUUCCUUAGCGGCCGCAGUGAAAAAUAUUAAAUUUUACCUCCACACUGGGGC .--.(((((.....)))))...(((((.((((.....))))....((((...........................((....)).((((((.........))))))))))....))))). ( -26.70) >DroSim_CAF1 6192 118 + 1 A--ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGGGCACAAAAGACAAAACAGAAAUUCCUUAGUGGCCGCAGUGAAAAAUAUUAAAUUUUACCUCCACACUGGGGC .--.((((........(((....)))..((((.....))))....))))......................((((((((......((((((.........)))))).....)))))))). ( -28.60) >DroEre_CAF1 5106 117 + 1 UAUACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGGGC--AAAAGACAAUACACAAAUUCCUUACCAGCCGCAGUGAAAAAUAUUAAAUUUUACCUCCGCAC-GGGGG ....(((((.....)))))...((((..((((((((((((((....))).--))))))..........)))))......((.(..((((((.........))))))..).))..-)))). ( -27.90) >DroYak_CAF1 5708 120 + 1 AAUACCCCCACAAUUGGGGAAAUCCUACGGAAUUCUUUUCCUUUCGGGGCACAAAAGACAAUACCAAAAUUCCUUACCAGCCACAGUGAAAAAUAUUAAAUUUUACCUCCACACAGGGGG ....(((((.....(((((.........((((((((((((((....)))...))))))..........)))))............((((((.........)))))))))))....))))) ( -24.30) >consensus A__ACCCCAACAAUUGGGGAAAUCCCACGGAAUUCUUUUCCUUUCGGGGCACAAAAGACAACACAGAAAUUCCUUAGCGGCCGCAGUGAAAAAUAUUAAAUUUUACCUCCACACUGGGGC ....(((((.....(((((......(((((((.....))))....(.(((....(((...............)))....))).).))).(((((.....)))))..)))))...))))). (-22.22 = -22.18 + -0.04)

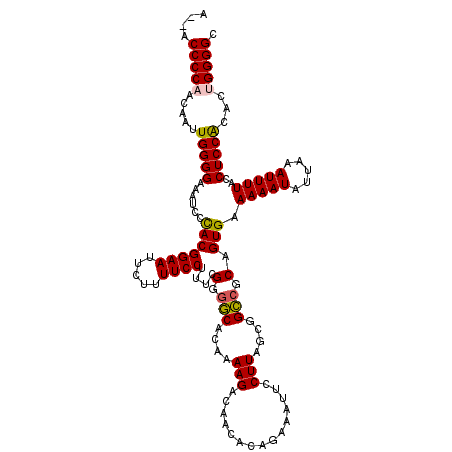
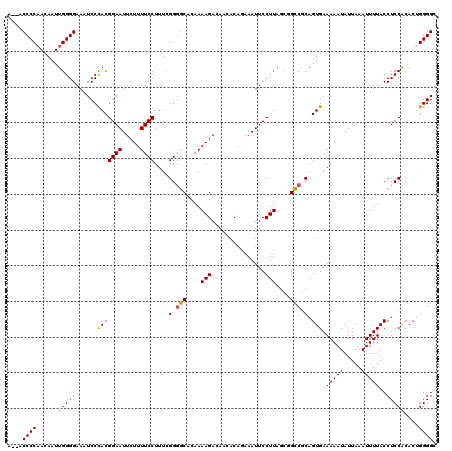
| Location | 19,312,828 – 19,312,946 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.29 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19312828 118 - 22407834 GCCCCAGUGUGGAGGUAAAAUUUAAUAUUUUUCACUGCGUCCGCUAAGGAAUUUCUGUGUUGUCUUUUGUGACCCAAAAGGAAAAGAAUUCCGUGGGAUUUCCCCAAUUGUUGGGGU--U (((((((.(((((.(((..................))).)))))...((((((.((......(((((((.....)))))))...)))))))).((((.....))))....)))))))--. ( -35.67) >DroSec_CAF1 5709 118 - 1 GCCCCAGUGUGGAGGUAAAAUUUAAUAUUUUUCACUGCGGCCGCUAAGGAAUUUCUGUGUUGUCUUUUGUGCUCCGAAAGGAAAAGAAUUCCGUGGGAUUUCCCCAAUUGUUGGGGU--U (((.(((((.(((((((........)))))))))))).)))..(((.(((((((..(..(........)..)(((....)))...))))))).))).....(((((.....))))).--. ( -35.20) >DroSim_CAF1 6192 118 - 1 GCCCCAGUGUGGAGGUAAAAUUUAAUAUUUUUCACUGCGGCCACUAAGGAAUUUCUGUUUUGUCUUUUGUGCCCCGAAAGGAAAAGAAUUCCGUGGGAUUUCCCCAAUUGUUGGGGU--U (((.(((((.(((((((........)))))))))))).)))(((.(((((............))))).)))((((((..((((.....)))).((((.....))))....)))))).--. ( -34.60) >DroEre_CAF1 5106 117 - 1 CCCCC-GUGCGGAGGUAAAAUUUAAUAUUUUUCACUGCGGCUGGUAAGGAAUUUGUGUAUUGUCUUUU--GCCCCGAAAGGAAAAGAAUUCCGUGGGAUUUCCCCAAUUGUUGGGGUAUA .((((-(.((((((((((((..((((((..(((..(((.....)))..))).....))))))..))))--)))((....))........)))))(((.....)))......))))).... ( -36.30) >DroYak_CAF1 5708 120 - 1 CCCCCUGUGUGGAGGUAAAAUUUAAUAUUUUUCACUGUGGCUGGUAAGGAAUUUUGGUAUUGUCUUUUGUGCCCCGAAAGGAAAAGAAUUCCGUAGGAUUUCCCCAAUUGUGGGGGUAUU (((((...(((((((.............))))))).(..(.(((...(((((((((((((........)))))((....))...))))))))...((....))))).)..)))))).... ( -34.62) >consensus GCCCCAGUGUGGAGGUAAAAUUUAAUAUUUUUCACUGCGGCCGCUAAGGAAUUUCUGUAUUGUCUUUUGUGCCCCGAAAGGAAAAGAAUUCCGUGGGAUUUCCCCAAUUGUUGGGGU__U .((((((.((((.(((((((........)))).)))....))))...(((((((........(((((((.....)))))))....))))))).((((.....))))....)))))).... (-26.06 = -26.10 + 0.04)

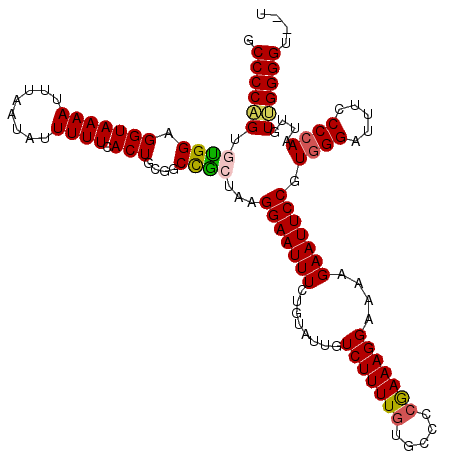
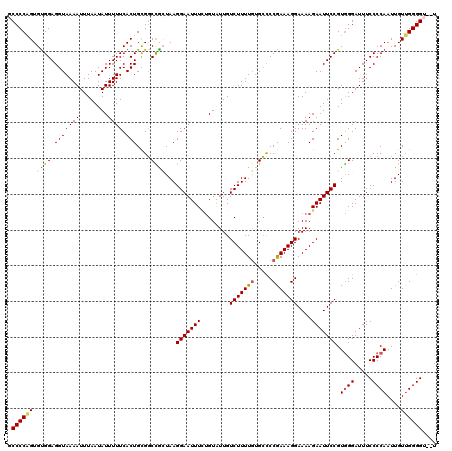
| Location | 19,312,946 – 19,313,062 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19312946 116 - 22407834 UGUUGGAGAUGCCUCUCCAGCCCCCAAAGAUCCCAUUGGGUUUUUGCGUUUCUUUU----UUUUUUUUUGAAGAUCCCAGUUCCCAACCGCAUUUUUUGGCUAUUUGUGAACGAUUAUUU .((((((((....))))))))..(((((((.....(((((...(((.(..(((((.----.........)))))..))))..)))))......)))))))....(((....)))...... ( -29.10) >DroSec_CAF1 5827 119 - 1 UGUUGGAGAUGCCUCUCCAGCCCCCAAAGAUCCCAUUGGGUUUUUGCGUUUUUUUUUCUUUUUUUUUUUGAAGAUCCCAGUUCCCAACCGCAUUU-UUGGCUAUUUAUGAACGAUUAUUU .((((((((....))))))))..((((((((.(..(((((...(((.(..(((((..............)))))..))))..)))))..).))))-)))).................... ( -27.14) >DroSim_CAF1 6310 109 - 1 UGUUGGAGAUGCCUCUCCAGCCCCCAAAGAUCCCAUUGGGUUUUUGCGUUUUU---------UUUUUUUGAAGAUC-CAGUUCCCAACCGCAUUU-UUGGCUAUUUAUGAACGAUUAUUU .((((((((....))))))))..((((((((.(..(((((...(((.((((((---------.......)))))).-)))..)))))..).))))-)))).................... ( -28.70) >DroEre_CAF1 5223 100 - 1 UGUUGGAGAUGCCUCUCCAGCC-CCAAAGAUCCCAUUGGGUUUUUGUGUU-----------------UUGGAGUUCCCAGUUC-CAACCGCAUUU-UCGCCUAUUUAUGAAAAAUUAUUU .(((((((.((...((((((..-((((((((((....))))))))).)..-----------------))))))....)).)))-))))....(((-(((........))))))....... ( -28.90) >DroYak_CAF1 5828 102 - 1 UGUUGAAGAUGCCUCUCAAGCCCCCAAAGAUCCCAUUGGGUUUUUGUGUU-----------------UUGGAGUUCCCAGUUCCCAACCGCAUUU-UUGCCUAUUUAUGAAAAAUUAUCU .(..((((((((((((.((((...(((((((((....))))))))).)))-----------------).))))......((.....)).))))))-))..)................... ( -20.20) >consensus UGUUGGAGAUGCCUCUCCAGCCCCCAAAGAUCCCAUUGGGUUUUUGCGUUU_U_________UUUUUUUGAAGAUCCCAGUUCCCAACCGCAUUU_UUGGCUAUUUAUGAACGAUUAUUU .((((((((....))))))))...(((((((((....))))))))).......................................................................... (-16.92 = -17.32 + 0.40)

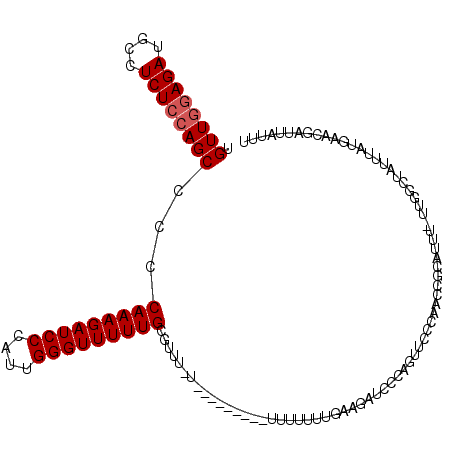
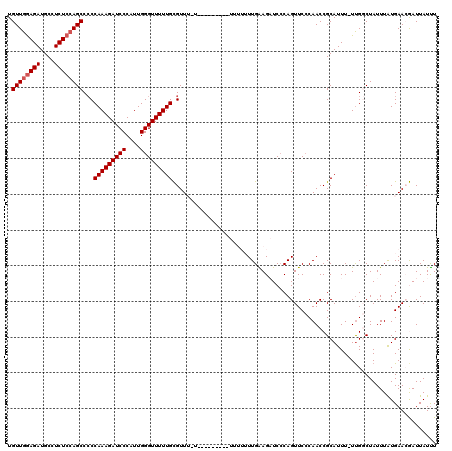
| Location | 19,313,022 – 19,313,142 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -39.60 |
| Energy contribution | -39.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19313022 120 + 22407834 CCCAAUGGGAUCUUUGGGGGCUGGAGAGGCAUCUCCAACAGCUGCUGGUAAUUAAUGUCUGGCCGCGUUCAAUGCACUGGAUAAUCACGCGGCUGGGGCCACAACUCUUUCCCCGAAAAU (((...)))...((((((((.((((((....)))))).(((((((((((......((((..(..((((...)))).)..)))))))).)))))))(((......)))..))))))))... ( -42.80) >DroSec_CAF1 5906 120 + 1 CCCAAUGGGAUCUUUGGGGGCUGGAGAGGCAUCUCCAACAGCUGCUGGUAAUUAAUGUCUGGCCGCGUUCAAUGCACUGGAUAAUCACGCGGCUGGGGCCACAACUCUUUCCCCGAAAAU (((...)))...((((((((.((((((....)))))).(((((((((((......((((..(..((((...)))).)..)))))))).)))))))(((......)))..))))))))... ( -42.80) >DroSim_CAF1 6379 119 + 1 CCCAAUGGGAUCUUUGGGGGCUGGAGAGGCAUCUCCAACAGCUGCUGGUAAUUAAUGUCUGGCCGCGUUCAAUGCACUGGAUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC-AAAAU (((...)))...((((((((.((((((....)))))).(((((((((((......((((..(..((((...)))).)..)))))))).)))))))(((......)))..)))))-))).. ( -43.10) >DroEre_CAF1 5284 118 + 1 CCCAAUGGGAUCUUUGG-GGCUGGAGAGGCAUCUCCAACAGCUGCUGGUAAUUAAUGUCUGGCCGCGUUCAAUGCACUGGAUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC-AAAAU ......((((...(((.-(((((((((....)))))..(((((((((((......((((..(..((((...)))).)..)))))))).))))))).)))).))).....)))).-..... ( -42.10) >DroYak_CAF1 5890 119 + 1 CCCAAUGGGAUCUUUGGGGGCUUGAGAGGCAUCUUCAACAGCUGCUGGUAAUUAAUGUCUGGCCGCGUUCAAUGCACUGGAUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC-AAAAU (((...)))...((((((((...(((((.......(..(((((((((((......((((..(..((((...)))).)..)))))))).)))))))..)......))))))))))-))).. ( -40.52) >consensus CCCAAUGGGAUCUUUGGGGGCUGGAGAGGCAUCUCCAACAGCUGCUGGUAAUUAAUGUCUGGCCGCGUUCAAUGCACUGGAUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC_AAAAU ......((((...(((..(((((((((....)))))..(((((((((((......((((..(..((((...)))).)..)))))))).))))))).)))).))).....))))....... (-39.60 = -39.64 + 0.04)

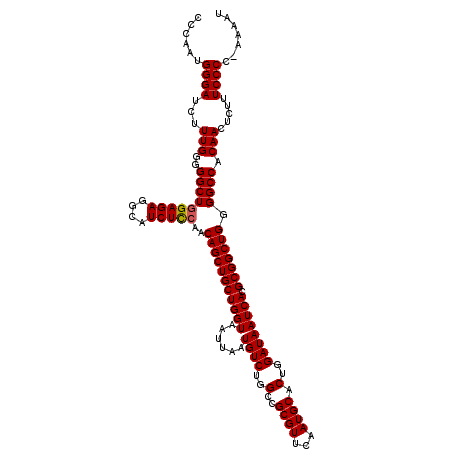
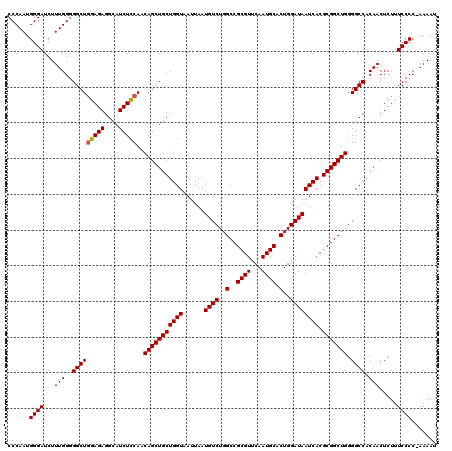
| Location | 19,313,102 – 19,313,222 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.74 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -33.06 |
| Energy contribution | -33.06 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19313102 120 + 22407834 AUAAUCACGCGGCUGGGGCCACAACUCUUUCCCCGAAAAUCUCGGCAAAUAAAUCUCUGACGAGGGUGGAACAGAGUUAUUCCUCCUUAGACUCCAUAUAAAUGAUAGUCCUGGCUGCAG ........(((((..((((...((((((((((((((.....))))........((((....))))..)))).))))))................(((....)))...))))..))))).. ( -35.90) >DroSec_CAF1 5986 120 + 1 AUAAUCACGCGGCUGGGGCCACAACUCUUUCCCCGAAAAUCUCGGCAAAUAAAUCUCUGACGAGGGUGGAACAGAGUUAUUCCUCCUAAGACUCCAUAUAAAUGAUAGUCCUGGCUGCAG ........(((((..((((...((((((((((((((.....))))........((((....))))..)))).))))))................(((....)))...))))..))))).. ( -35.90) >DroSim_CAF1 6459 119 + 1 AUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC-AAAAUCUCGGCAAAUAAAUCUCUGACGAGGGUGGAACAGAGUUAUUCCUCCUUAGACUCCAAAUAAAUGAUAGUCCUGGCUGCAG ........(((((..((((...((((((((((((-....((((((...........)))).)).)).)))).))))))................((......))...))))..))))).. ( -34.00) >DroEre_CAF1 5363 119 + 1 AUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC-AAAAUCUCGGCAAAUAAAUCUCUGACGAGGGUGGAACAGAGUUAUUCCUCCUCAGACUCAAUAUAAAUGAUAGUCCUGGCUGCAG ........(((((..((((...((((((((((((-....((((((...........)))).)).)).)))).))))))..............(((.......)))..))))..))))).. ( -34.70) >DroYak_CAF1 5970 119 + 1 AUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC-AAAAUCUCGGCAAAUAAAUCUCUGACGAGGGUGGAACAGAGUUAUUCCUCCUUAGACUCAGUAUAAAUGAUAGUCCUGGCUGCAG ........(((((..((((...((((((((((((-....((((((...........)))).)).)).)))).))))))..............(((.......)))..))))..))))).. ( -35.50) >consensus AUAAUCACGCGGCUGGGGCCACAACUCUUUCCCC_AAAAUCUCGGCAAAUAAAUCUCUGACGAGGGUGGAACAGAGUUAUUCCUCCUUAGACUCCAUAUAAAUGAUAGUCCUGGCUGCAG ........(((((..((((...((((((((((((......((((.((..........)).)))))).)))).))))))....((....)).................))))..))))).. (-33.06 = -33.06 + -0.00)

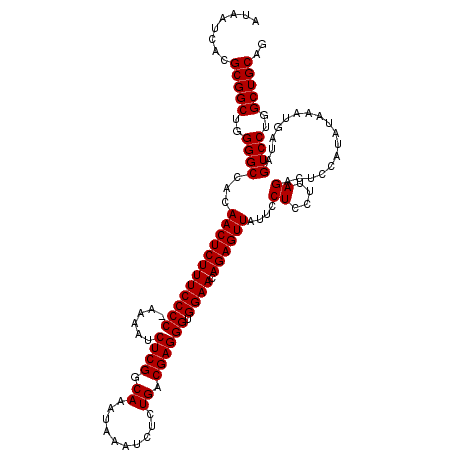
| Location | 19,313,102 – 19,313,222 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.74 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -35.44 |
| Energy contribution | -36.24 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19313102 120 - 22407834 CUGCAGCCAGGACUAUCAUUUAUAUGGAGUCUAAGGAGGAAUAACUCUGUUCCACCCUCGUCAGAGAUUUAUUUGCCGAGAUUUUCGGGGAAAGAGUUGUGGCCCCAGCCGCGUGAUUAU ..((.((..(((((.((((....)))))))))..((.((.((((((((.((((.((((((.((((......)))).))))......))))))))))))))..)))).)).))........ ( -38.40) >DroSec_CAF1 5986 120 - 1 CUGCAGCCAGGACUAUCAUUUAUAUGGAGUCUUAGGAGGAAUAACUCUGUUCCACCCUCGUCAGAGAUUUAUUUGCCGAGAUUUUCGGGGAAAGAGUUGUGGCCCCAGCCGCGUGAUUAU ..((.((.((((((.((((....)))))))))).((.((.((((((((.((((.((((((.((((......)))).))))......))))))))))))))..)))).)).))........ ( -39.00) >DroSim_CAF1 6459 119 - 1 CUGCAGCCAGGACUAUCAUUUAUUUGGAGUCUAAGGAGGAAUAACUCUGUUCCACCCUCGUCAGAGAUUUAUUUGCCGAGAUUUU-GGGGAAAGAGUUGUGGCCCCAGCCGCGUGAUUAU ..((.((..(((((.(((......))))))))..((.((.((((((((.((((.((((((.((((......)))).)))).....-))))))))))))))..)))).)).))........ ( -38.20) >DroEre_CAF1 5363 119 - 1 CUGCAGCCAGGACUAUCAUUUAUAUUGAGUCUGAGGAGGAAUAACUCUGUUCCACCCUCGUCAGAGAUUUAUUUGCCGAGAUUUU-GGGGAAAGAGUUGUGGCCCCAGCCGCGUGAUUAU ..((.((((((....(((.......))).)))).((.((.((((((((.((((.((((((.((((......)))).)))).....-))))))))))))))..)))).)).))........ ( -36.40) >DroYak_CAF1 5970 119 - 1 CUGCAGCCAGGACUAUCAUUUAUACUGAGUCUAAGGAGGAAUAACUCUGUUCCACCCUCGUCAGAGAUUUAUUUGCCGAGAUUUU-GGGGAAAGAGUUGUGGCCCCAGCCGCGUGAUUAU ..((.((..(((((.............)))))..((.((.((((((((.((((.((((((.((((......)))).)))).....-))))))))))))))..)))).)).))........ ( -35.22) >consensus CUGCAGCCAGGACUAUCAUUUAUAUGGAGUCUAAGGAGGAAUAACUCUGUUCCACCCUCGUCAGAGAUUUAUUUGCCGAGAUUUU_GGGGAAAGAGUUGUGGCCCCAGCCGCGUGAUUAU ..((.((..(((((.((((....)))))))))..((.((.((((((((.((((.((((((.((((......)))).))))......))))))))))))))..)))).)).))........ (-35.44 = -36.24 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:36 2006