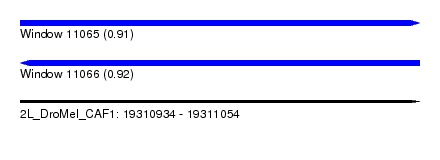
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,310,934 – 19,311,054 |
| Length | 120 |
| Max. P | 0.919885 |

| Location | 19,310,934 – 19,311,054 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
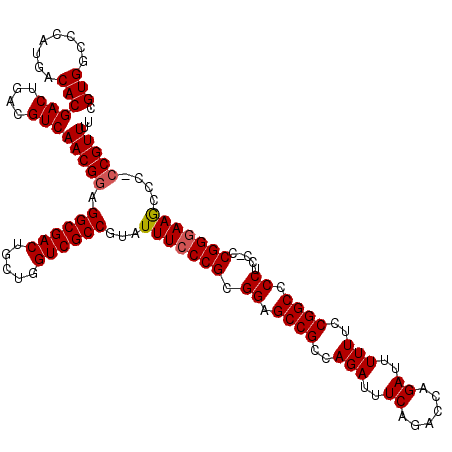
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -50.08 |
| Consensus MFE | -44.00 |
| Energy contribution | -43.44 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19310934 120 + 22407834 AAAACGGUGGGCUUUCCGGGGGAGGGGCCGGAAAAAAUCUGGUCUGAAAUCUGGCGGCUCCGCGGGAAAUACGGCGACCAGCAGUCGCCGUCGUUGACGUCAGUCGUGUCAUGGGCCACG ......((((.((((((.(.((((..((((((.....((......))..))))))..)))).).))))..((((((((.....))))))))...(((((.(....)))))).)).)))). ( -48.50) >DroSec_CAF1 3776 119 + 1 AAAACGG-GGGCUUCCCGGGGACGGGGCCGGAAAAAAUCUGGUCUGAAAUCUGGCGGCUCCGCGGGAAAUACGGCGACCAGCAGUCGCCGCCGUUGACGUCAGUCGUGUCAUGGGCCACG ....(((-(((((.((((....))))((((((.....((......))..)))))))))))).))((.....(((((((.....)))))))((((.((((.(....))))))))).))... ( -50.40) >DroSim_CAF1 4247 120 + 1 AAAACGGCGGGCUUCCCGGCGGAGGGGCCGGAAAAAAUCUGGUCUGAAAUCUGGCGGCUCCGCGGGAAAUACGGCGACCAGCAGUCGCCACCGUUGACGUCAGUCGUGUCAUGGGCCACG .....(((....((((((.(((((..((((((.....((......))..))))))..)))))))))))....((((((.....)))))).((((.((((.(....))))))))))))... ( -51.50) >DroEre_CAF1 3247 116 + 1 AAAACGG--GGUUUCCCGG--GAGGGGCCGGAAAAAAUCUGGUCUGAAAUCUGGCGGCUCCGCGGGAAAUACGGCGACCAGCAGUCGCCUCCGUUGACGUCAGUCGUGUCAUGGGCCACG ..(((((--((((((((((--(((..((((((.....((......))..))))))..)))).))))))))..((((((.....))))))))))))(((....)))(((........))). ( -50.00) >DroYak_CAF1 3811 116 + 1 AAAACGG--GGUUUCCCGG--GAGGGGCCGGAAAAAAUCUGGUCUGAAAUCUGGCGGCUCCGCGGGAAAUACGGCGACCAACAGUCGCCUCCGUUGACGUCAGUCGUGUCAUGGGCCACG ..(((((--((((((((((--(((..((((((.....((......))..))))))..)))).))))))))..((((((.....))))))))))))(((....)))(((........))). ( -50.00) >consensus AAAACGG_GGGCUUCCCGG_GGAGGGGCCGGAAAAAAUCUGGUCUGAAAUCUGGCGGCUCCGCGGGAAAUACGGCGACCAGCAGUCGCCGCCGUUGACGUCAGUCGUGUCAUGGGCCACG ....((...((((((((.......((((((((.....))))))))........(((....))))))))....((((((.....)))))).((((.((((.(....)))))))))))).)) (-44.00 = -43.44 + -0.56)

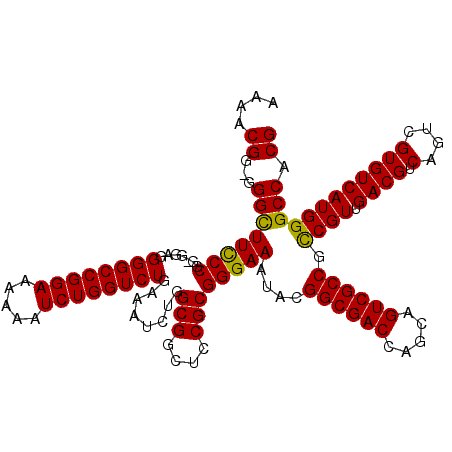
| Location | 19,310,934 – 19,311,054 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -45.66 |
| Consensus MFE | -39.34 |
| Energy contribution | -39.50 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19310934 120 - 22407834 CGUGGCCCAUGACACGACUGACGUCAACGACGGCGACUGCUGGUCGCCGUAUUUCCCGCGGAGCCGCCAGAUUUCAGACCAGAUUUUUUCCGGCCCCUCCCCCGGAAAGCCCACCGUUUU .((((............((((.(((....((((((((.....)))))))).......(((....)))..))).))))........((((((((........)))))))).))))...... ( -40.80) >DroSec_CAF1 3776 119 - 1 CGUGGCCCAUGACACGACUGACGUCAACGGCGGCGACUGCUGGUCGCCGUAUUUCCCGCGGAGCCGCCAGAUUUCAGACCAGAUUUUUUCCGGCCCCGUCCCCGGGAAGCCC-CCGUUUU .(((........)))(((....)))((((((((((((.....)))))))..((((((((((.((((..(((..((......))..)))..)))).)))....)))))))...-))))).. ( -45.40) >DroSim_CAF1 4247 120 - 1 CGUGGCCCAUGACACGACUGACGUCAACGGUGGCGACUGCUGGUCGCCGUAUUUCCCGCGGAGCCGCCAGAUUUCAGACCAGAUUUUUUCCGGCCCCUCCGCCGGGAAGCCCGCCGUUUU .(((........)))(((....)))((((((((((((.....))))))(..((((((((((((..(((.((..((......)).....)).)))..))))).)))))))..))))))).. ( -50.50) >DroEre_CAF1 3247 116 - 1 CGUGGCCCAUGACACGACUGACGUCAACGGAGGCGACUGCUGGUCGCCGUAUUUCCCGCGGAGCCGCCAGAUUUCAGACCAGAUUUUUUCCGGCCCCUC--CCGGGAAACC--CCGUUUU .(((........)))(((....)))(((((.((((((.....))))))...(((((((.((((..(((.((..((......)).....)).)))..)))--))))))))..--))))).. ( -45.80) >DroYak_CAF1 3811 116 - 1 CGUGGCCCAUGACACGACUGACGUCAACGGAGGCGACUGUUGGUCGCCGUAUUUCCCGCGGAGCCGCCAGAUUUCAGACCAGAUUUUUUCCGGCCCCUC--CCGGGAAACC--CCGUUUU .(((........)))(((....)))(((((.((((((.....))))))...(((((((.((((..(((.((..((......)).....)).)))..)))--))))))))..--))))).. ( -45.80) >consensus CGUGGCCCAUGACACGACUGACGUCAACGGAGGCGACUGCUGGUCGCCGUAUUUCCCGCGGAGCCGCCAGAUUUCAGACCAGAUUUUUUCCGGCCCCUCC_CCGGGAAGCCC_CCGUUUU .(((........)))(((....)))(((((.((((((.....))))))...(((((((.((.((((..(((..((......))..)))..)))).)).....)))))))....))))).. (-39.34 = -39.50 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:27 2006