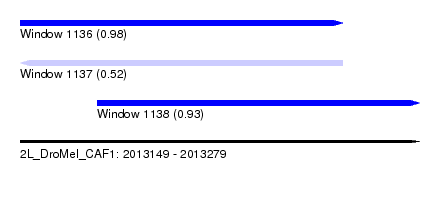
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,013,149 – 2,013,279 |
| Length | 130 |
| Max. P | 0.977381 |

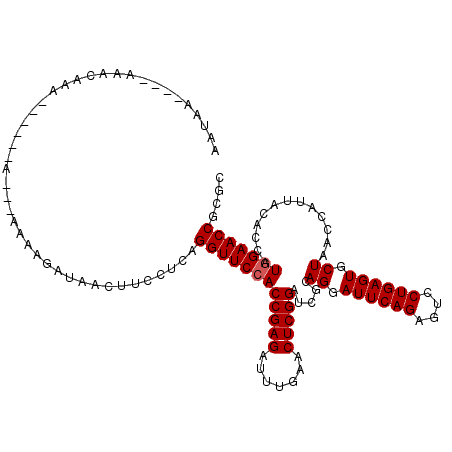
| Location | 2,013,149 – 2,013,254 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.57 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2013149 105 + 22407834 AACAA----AAACAAA------A---AAAAAAGGAAUUCCUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCGC .....----.......------.---.....(((....)))..((((((((((((.......))))).....((.((((((....)))))).)).............))))))).... ( -28.70) >DroGri_CAF1 279087 99 + 1 UACAA----AAAUAAU-CGGUAUAAA--------------UCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCCGG .....----.......-(((......--------------...((((((((((((.......))))).....((.((((((....)))))).)).............)))))))))). ( -27.30) >DroSec_CAF1 30703 99 + 1 ---------AUAUAAA------A---AA-AAUUGAUUCUCUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCGC ---------.......------.---..-..............((((((((((((.......))))).....((.((((((....)))))).)).............))))))).... ( -26.90) >DroEre_CAF1 27864 110 + 1 AAUAA----AAACAAA-AACGAA---AAAAGAUAACUUCUUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCGC .....----.......-......---..((((.....))))..((((((((((((.......))))).....((.((((((....)))))).)).............))))))).... ( -27.70) >DroYak_CAF1 32230 115 + 1 AAUAAAUAAAAACAGACAAAAAA---AAAAGACAACUUUCUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCGCGG .......................---.................(((((..(((((.......))))).....((.((((((....)))))).))...............))))).... ( -22.20) >DroPer_CAF1 34911 99 + 1 --UAA----AAAUAAA------A----AAAGAUAAC--GUUAAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-CAC --...----.......------.----.........--.....((((((((((((.......))))).....((.((((((....)))))).)).............)))))))-... ( -26.00) >consensus AAUAA____AAACAAA______A___AAAAGAUAACUUCCUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCGC ...........................................((((((((((((.......))))).....((.((((((....)))))).)).............))))))).... (-25.40 = -25.57 + 0.17)

| Location | 2,013,149 – 2,013,254 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.22 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2013149 105 - 22407834 GCGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGAGGAAUUCCUUUUUU---U------UUUGUUU----UUGUU ....(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).((((....))))....---.------.......----..... ( -28.70) >DroGri_CAF1 279087 99 - 1 CCGGGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGA--------------UUUAUACCG-AUUAUUU----UUGUA ...((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..--------------.........-.......----..... ( -26.90) >DroSec_CAF1 30703 99 - 1 GCGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGAGAGAAUCAAUU-UU---U------UUUAUAU--------- ....(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))((((((((.....)-))---)------))))...--------- ( -28.40) >DroEre_CAF1 27864 110 - 1 GCGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGAAGAAGUUAUCUUUU---UUCGUU-UUUGUUU----UUAUU (((.(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).(((((((....)))))---))))).-.......----..... ( -28.90) >DroYak_CAF1 32230 115 - 1 CCGCGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGAGAAAGUUGUCUUUU---UUUUUUGUCUGUUUUUAUUUAUU ....(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).(((((((....)))))---))..................... ( -26.20) >DroPer_CAF1 34911 99 - 1 GUG-GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUUAAC--GUUAUCUUU----U------UUUAUUU----UUA-- (((-(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..))--.........----.------.......----...-- ( -26.40) >consensus GCGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGAGAAAGUUAUCUUUU___U______UUUAUUU____UUAUU ....(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))........................................... (-25.35 = -25.22 + -0.14)

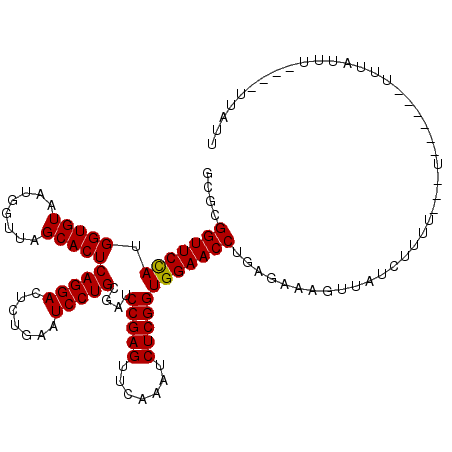
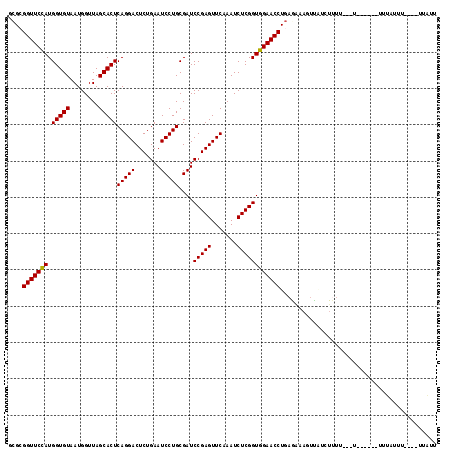
| Location | 2,013,174 – 2,013,279 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.03 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2013174 105 + 22407834 CCUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCGCUGAUCA----CCCGUCGCCAAA---UC--------GUUCA .....((((((((((((.......))))).....((.((((((....)))))).)).............)))))))(((.((....----..)).)))....---..--------..... ( -30.20) >DroVir_CAF1 22133 116 + 1 GCUCAGGUUCCACCGAGACUUGAACUCGGAUCGCAGGAUUCAGAGUCCUAAGUGCUAACCAUUACACCAUGGAACCUGUUAGAUAA----GCGACCACAAACAAACAAAAAUUUUGCCGU (((((((((((((((((.......)))))..((((((((.....)))))..)))...............))))))))).......)----))............................ ( -28.51) >DroPse_CAF1 36096 104 + 1 GUUAAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-CACUGGGCG----AGGGCCAUCAAA---GA--------GCUCU .....(((((..(((((.......))))).((((((.((((((....)))))).))..........((((((...)-)).))))))----))))))......---..--------..... ( -31.30) >DroGri_CAF1 279108 118 + 1 --UCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCCGGUGAUCAAGAAUCGACAGCAAACAAGCAAAAUUUUAUUACU --...((((((((((((.......))))).....((.((((((....)))))).)).............))))))).((((........))))...((......)).............. ( -29.90) >DroMoj_CAF1 21196 115 + 1 CC-UAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCAACUGAUCA----GUGAUCGCCGAGAAACAAAACUUGAAUUCU ..-..((((((((((((.......))))).....((.((((((....)))))).)).............))))))).....((((.----..))))..((((........))))...... ( -29.60) >DroAna_CAF1 28086 105 + 1 UCCAAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCUAUGGUCG----GUCACUGCCAGA---UC--------GUUCC .....((((((((((((.......))))).....((.((((((....)))))).)).............)))))))...(((((((----((....))).))---))--------))... ( -33.60) >consensus CCUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCCACUGAUCA____GCGACCGCCAAA___CA________GUUCU .....((((((((((((.......))))).....((.((((((....)))))).)).............)))))))............................................ (-25.32 = -25.48 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:08 2006