
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
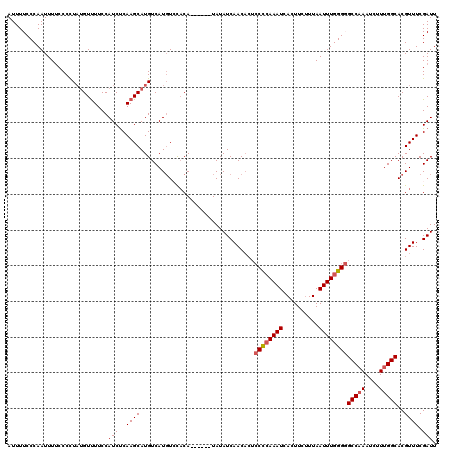
| Location | 19,308,122 – 19,308,306 |
| Length | 184 |
| Max. P | 0.926167 |

| Location | 19,308,122 – 19,308,236 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.71 |
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19308122 114 + 22407834 AUUUUCCCAAUUUUCCCCUAAGUUUUCCAUCUCAAGCAUGUCAUGUCCACA------UAUAUCAACACUCCCCAAAUCACUUCCUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUU ...............................((((((((((.(((....))------))))).......((((((((..........))))))))(((((....)))))..)))).)).. ( -18.40) >DroSec_CAF1 819 114 + 1 AUUUUCCCAAUUUUCCCCUAUGUUUUCCAUCUCAAGCAUGUCAUGUCCACA------UAUAUCAACACUCCCAAAAUCACUUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUU ..............(((((((((((........)))))))..(((....))------).................................))))(((((....)))))........... ( -16.90) >DroSim_CAF1 754 114 + 1 AUUUUCCCAAUUUUCCCCUAUGUUUUCCAUCUCAAGCAUGUCAUGUCCACA------UAUAUCAACACUCCCCAAAUCACUUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUU ..................(((((((........)))))))...........------...(((......((((((((..........))))))))(((((....))))).......))). ( -19.70) >DroEre_CAF1 658 115 + 1 AUUUUCCCAAUUUCCCCUU-UGUUUUCCAUCCCAAGCAUGUCAUGUGUCCACACAAGUAUAUCAACU---CCCAAAUCACUUCUUUAAUUUGGG-GCCCAAUCUUUGGCACGUUUCGAUU ...................-...........(.((((.(((((((((....))))...........(---(((((((..........)))))))-).........))))).)))).)... ( -17.30) >DroYak_CAF1 894 113 + 1 AUUUUCCCAAUUUUCCCUUAUGUCUUCCAUCUCAAGCAUGUCAUUUGUACA------UAUAUCAACACUCCUCAAAUCACUUCUUUAAUUUGGG-GCCAAAUCUUUGGCACGUUUCGAUU .....(((((.........(((.(((.......))))))((.(((((....------...............))))).)).........)))))-(((((....)))))........... ( -14.21) >consensus AUUUUCCCAAUUUUCCCCUAUGUUUUCCAUCUCAAGCAUGUCAUGUCCACA______UAUAUCAACACUCCCCAAAUCACUUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUU ............................(((..((((................................((((((((..........))))))))(((((....)))))..)))).))). (-13.23 = -13.72 + 0.49)


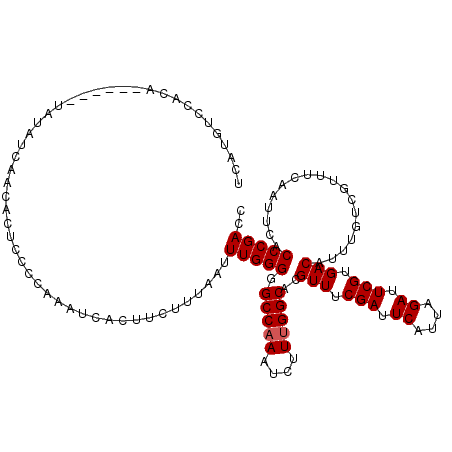
| Location | 19,308,162 – 19,308,276 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19308162 114 + 22407834 UCAUGUCCACA------UAUAUCAACACUCCCCAAAUCACUUCCUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACC ..(((((.((.------......(((...((((((((..........))))))))(((((....)))))..)))..((.((....)).)))))))))..((((............)))). ( -25.70) >DroSec_CAF1 859 114 + 1 UCAUGUCCACA------UAUAUCAACACUCCCAAAAUCACUUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACC ..(((((.((.------......(((..(((((((.............)))))))(((((....)))))..)))..((.((....)).)))))))))..((((............)))). ( -21.62) >DroSim_CAF1 794 114 + 1 UCAUGUCCACA------UAUAUCAACACUCCCCAAAUCACUUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACC ..(((((.((.------......(((...((((((((..........))))))))(((((....)))))..)))..((.((....)).)))))))))..((((............)))). ( -25.70) >DroEre_CAF1 697 116 + 1 UCAUGUGUCCACACAAGUAUAUCAACU---CCCAAAUCACUUCUUUAAUUUGGG-GCCCAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAGUUCACCCGAAC ((..(((..(((((((((.........---(((((((..........)))))))-(((........)))..(((.(((.((....)).))).))))))))).))....)..)))..)).. ( -21.70) >DroYak_CAF1 934 113 + 1 UCAUUUGUACA------UAUAUCAACACUCCUCAAAUCACUUCUUUAAUUUGGG-GCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGAAC ...........------...............................((((((-(((((....)))))..(((.(((.((....)).))).)))..................)))))). ( -20.10) >consensus UCAUGUCCACA______UAUAUCAACACUCCCCAAAUCACUUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACC .................................................(((((.(((((....)))))..(((.(((.((....)).))).)))..................))))).. (-17.90 = -18.10 + 0.20)


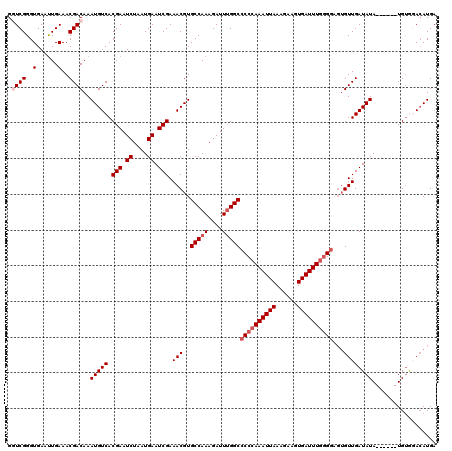
| Location | 19,308,162 – 19,308,276 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -23.13 |
| Energy contribution | -24.38 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19308162 114 - 22407834 GGUCGGGUGAAUUGAAACGACAAAUGUCACGAAUCUAAUGAAUCGAAACGUGCCAAAGAUUUGGCCCCCAAAUUAAGGAAGUGAUUUGGGGAGUGUUGAUAUA------UGUGGACAUGA .((((..(......)..))))..(((((.(((.((....)).)))..(((((((((....)))))((((((((((......))))))))))...........)------))).))))).. ( -34.00) >DroSec_CAF1 859 114 - 1 GGUCGGGUGAAUUGAAACGACAAAUGUCACGAAUCUAAUGAAUCGAAACGUGCCAAAGAUUUGGCCCCCAAAUUAAAGAAGUGAUUUUGGGAGUGUUGAUAUA------UGUGGACAUGA .((((..(......)..))))..(((((.(((.((....)).)))..(((((((((....)))))(((.((((((......)))))).)))...........)------))).))))).. ( -27.50) >DroSim_CAF1 794 114 - 1 GGUCGGGUGAAUUGAAACGACAAAUGUCACGAAUCUAAUGAAUCGAAACGUGCCAAAGAUUUGGCCCCCAAAUUAAAGAAGUGAUUUGGGGAGUGUUGAUAUA------UGUGGACAUGA .((((..(......)..))))..(((((.(((.((....)).)))..(((((((((....)))))((((((((((......))))))))))...........)------))).))))).. ( -34.00) >DroEre_CAF1 697 116 - 1 GUUCGGGUGAACUGAAACGACAAAUGUCACGAAUCUAAUGAAUCGAAACGUGCCAAAGAUUGGGC-CCCAAAUUAAAGAAGUGAUUUGGG---AGUUGAUAUACUUGUGUGGACACAUGA .(((((.....))))).((((..((((..(((.((....)).)))..))))(((........)))-(((((((((......)))))))))---.)))).....(.((((....)))).). ( -29.90) >DroYak_CAF1 934 113 - 1 GUUCGGGUGAAUUGAAACGACAAAUGUCACGAAUCUAAUGAAUCGAAACGUGCCAAAGAUUUGGC-CCCAAAUUAAAGAAGUGAUUUGAGGAGUGUUGAUAUA------UGUACAAAUGA .(((((.....)))))...(((.(((((.(((.((....)).))).((((.(((((....)))))-(((((((((......))))))).))..))))))))).------)))........ ( -23.00) >consensus GGUCGGGUGAAUUGAAACGACAAAUGUCACGAAUCUAAUGAAUCGAAACGUGCCAAAGAUUUGGCCCCCAAAUUAAAGAAGUGAUUUGGGGAGUGUUGAUAUA______UGUGGACAUGA .((((..(......)..))))..(((((.(((.((....)).))).(((..(((((....)))))((((((((((......))))))))))...)))))))).................. (-23.13 = -24.38 + 1.25)


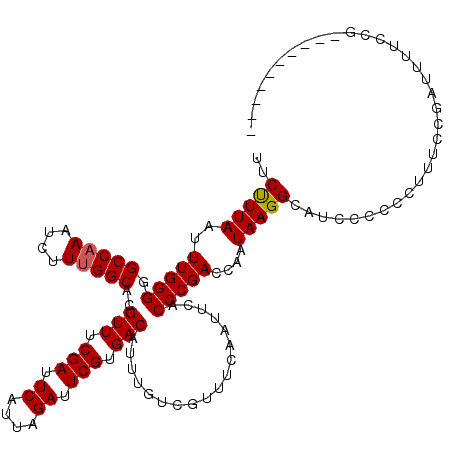
| Location | 19,308,196 – 19,308,306 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.98 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19308196 110 + 22407834 UUCCUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACCAAUAAGGCAUCCCCCCUUUCCGAUUUUCCG---------- ...........(((((((((....)))))..(((.(((.((....)).))).)))(((.((((............)))).))).........))))..............---------- ( -22.80) >DroSec_CAF1 893 108 + 1 UUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACCAAUAAGGCAUCCC-CCUUUCUGCUUUUCU----------- ...........(((((((((....)))))..(((.(((.((....)).))).)))(((.((((............)))).)))........))-)).............----------- ( -22.80) >DroSim_CAF1 828 119 + 1 UUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACCAAUAAGGCAUCCC-CCUUUCUGAUUUUCCGGUCCCCACCC ..........((((((((........((((.(((.(((.((....)).))).)))...)))).................((..((((......-))))..)).......))))))))... ( -28.90) >DroEre_CAF1 734 107 + 1 UUCUUUAAUUUGGG-GCCCAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAGUUCACCCGAACAAUAAGGUA-CCCCCUUUUCCGAUUUC-CG---------- ...........(((-(.((.......((((.(((.(((.((....)).))).)))...))))......((((....)))).....))..-.))))............-..---------- ( -21.00) >DroYak_CAF1 968 109 + 1 UUCUUUAAUUUGGG-GCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGAACAAUAAGGUAUCCCCCUUUUCCGAUUUCCCG---------- ........((((((-(((((....)))))..(((.(((.((....)).))).)))..................))))))....((((......)))).............---------- ( -21.80) >consensus UUCUUUAAUUUGGGGGCCAAAUCUUUGGCACGUUUCGAUUCAUUAGAUUCGUGACAUUUGUCGUUUCAAUUCACCCGACCAAUAAGGCAUCCCCCCUUUCCGAUUUUCCG__________ ..(((((..(((((.(((((....)))))..(((.(((.((....)).))).)))..................)))))....)))))................................. (-19.36 = -19.40 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:24 2006