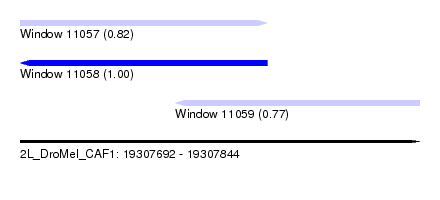
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,307,692 – 19,307,844 |
| Length | 152 |
| Max. P | 0.996537 |

| Location | 19,307,692 – 19,307,786 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19307692 94 + 22407834 AAAAGAAUGGCAGGCGGAAUUGCAAUGUCAGUGUCUCCGGGGAGGGGGAGCAAGCGGGUGAGAGCUGCACAAUGUUGCCACAUUGCGCCUGA---UG ..........((((((.(((.(((((((..((..((((.....))))..))..((((.(....)))))...)))))))...))).)))))).---.. ( -31.40) >DroSec_CAF1 361 97 + 1 AAAAGAAUGGCAGGCGGAAUUGCAAUGUCAGUGUCUCCGGGGAGGGGGAGCAACAGGGUGAGAGCUGCACAAUGUUGCCACAUUGCGCCUGAUGAUG ..........(((((......(((((((.....(((((......)))))((((((..(((.......)))..)))))).))))))))))))...... ( -32.50) >DroSim_CAF1 314 97 + 1 AAAAGAAUGGCAGGCGGAAUUGCAAUGUCAGUGUCUCCGGGGAGGGGGAGCAACAGGGUGAGAGCUGCACAAUGUUGCCACAUUGCGCCUGAUGAUG ..........(((((......(((((((.....(((((......)))))((((((..(((.......)))..)))))).))))))))))))...... ( -32.50) >DroEre_CAF1 162 97 + 1 AAAAGAAUGGCAGGCGGAAUUGCAAUGUCAGAGUCUCCGGGGAGGGGGAGCAUCAGAGGAGGAGCUGCACCAUGUUGCCACAUUGCGCCUGAUGAGG ..........((((((.(((.((((((((((..(((((...((..(....).))...)))))..))).))...)))))...))).))))))...... ( -31.80) >DroYak_CAF1 401 97 + 1 AAAAGAAUGGCAAGCGGAAUUGCAAUGUCAGUGUCUCCGGGGAGGGGGAGCAAAAGAAAAAAAACGGCGCAAUGUUGCAAGAUUGCGCCUGAUGAGG .........((((......))))...((((((..((((.....))))..))..............((((((((........)))))))))))).... ( -26.90) >consensus AAAAGAAUGGCAGGCGGAAUUGCAAUGUCAGUGUCUCCGGGGAGGGGGAGCAACAGGGUGAGAGCUGCACAAUGUUGCCACAUUGCGCCUGAUGAUG ..........((((((.(((.(((((((..((((((((.....)))).(((............))))))).)))))))...))).))))))...... (-23.70 = -24.38 + 0.68)

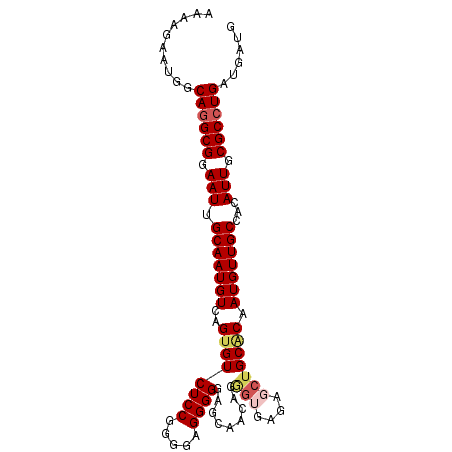
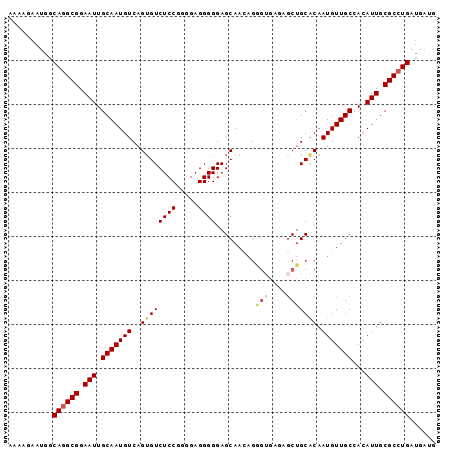
| Location | 19,307,692 – 19,307,786 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -21.00 |
| Energy contribution | -21.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19307692 94 - 22407834 CA---UCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUCACCCGCUUGCUCCCCCUCCCCGGAGACACUGACAUUGCAAUUCCGCCUGCCAUUCUUUU ..---.((((((((((((....)))))))(((((........)).((((((........)))).)).......)))......))))).......... ( -24.70) >DroSec_CAF1 361 97 - 1 CAUCAUCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUCACCCUGUUGCUCCCCCUCCCCGGAGACACUGACAUUGCAAUUCCGCCUGCCAUUCUUUU ......(((((((((((((((((((..(((.......)))..)))))))....(((....)))......)))))))......))))).......... ( -27.70) >DroSim_CAF1 314 97 - 1 CAUCAUCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUCACCCUGUUGCUCCCCCUCCCCGGAGACACUGACAUUGCAAUUCCGCCUGCCAUUCUUUU ......(((((((((((((((((((..(((.......)))..)))))))....(((....)))......)))))))......))))).......... ( -27.70) >DroEre_CAF1 162 97 - 1 CCUCAUCAGGCGCAAUGUGGCAACAUGGUGCAGCUCCUCCUCUGAUGCUCCCCCUCCCCGGAGACUCUGACAUUGCAAUUCCGCCUGCCAUUCUUUU ......((((((((..(((....)))..))).((......((.((..((((........))))..)).))....))......))))).......... ( -25.20) >DroYak_CAF1 401 97 - 1 CCUCAUCAGGCGCAAUCUUGCAACAUUGCGCCGUUUUUUUUCUUUUGCUCCCCCUCCCCGGAGACACUGACAUUGCAAUUCCGCUUGCCAUUCUUUU .....(((((((((((........)))))))).............((((((........)))).)).)))....((((......))))......... ( -20.70) >consensus CAUCAUCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUCACCCUGUUGCUCCCCCUCCCCGGAGACACUGACAUUGCAAUUCCGCCUGCCAUUCUUUU .....(((((((((((((....)))))))))................((((........))))...))))....(((........)))......... (-21.00 = -21.24 + 0.24)

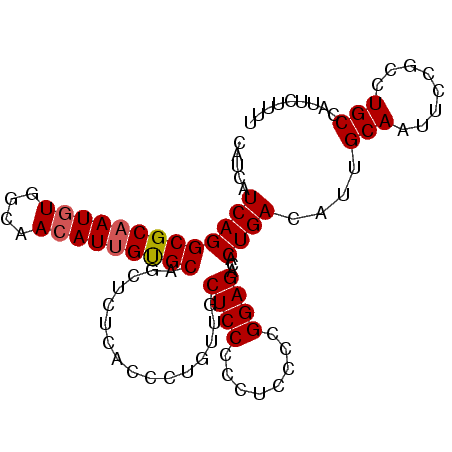
| Location | 19,307,751 – 19,307,844 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 71.97 |
| Mean single sequence MFE | -16.68 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19307751 93 - 22407834 CACUUACUCGCCGAAC-AAAGCUCGUGCCCCACCCCAAGCACCACCCCAACCUCCUGCUCA---UCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUC .........(((((..-.....))).)).........(((..............(((....---.)))(((((((((....))))))))).)))... ( -20.00) >DroSec_CAF1 420 95 - 1 CACUUACUCGCCCAAC-AAAGCCCGUGCCCCACC-CAAGCACCACCCCAACCUCCCGCUCAUCAUCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUC ................-..(((..((((......-...))))..........................(((((((((....))))))))).)))... ( -18.40) >DroSim_CAF1 373 82 - 1 CACUUACUCGCCCAAC-AAAGCCCG--------------CACCACCCCAACCUCCCGCUCAUCAUCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUC ................-..(((..(--------------(................))..........(((((((((....))))))))).)))... ( -14.39) >DroEre_CAF1 221 77 - 1 CACUUACUCGGCAA-C-AAAACCCGUGCCCCACC-CAA-----------GU------CCCCUCAUCAGGCGCAAUGUGGCAACAUGGUGCAGCUCCU .........((((.-(-.......))))).....-...-----------..------..........((((((..(((....)))..))).)))... ( -14.40) >DroYak_CAF1 460 88 - 1 CACUUACGCGCCCAGCAAAAACCCUCGCCCCACC-CAAGCCC--UCUCAGU------CCCCUCAUCAGGCGCAAUCUUGCAACAUUGCGCCGUUUUU .......((.....))((((((....((......-...))..--.......------..........((((((((........)))))))))))))) ( -16.20) >consensus CACUUACUCGCCCAAC_AAAGCCCGUGCCCCACC_CAAGCACCACCCCAACCUCC_GCUCAUCAUCAGGCGCAAUGUGGCAACAUUGUGCAGCUCUC ....................................................................(((((((((....)))))))))....... (-11.48 = -11.72 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:20 2006