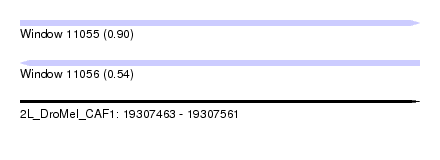
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,307,463 – 19,307,561 |
| Length | 98 |
| Max. P | 0.898179 |

| Location | 19,307,463 – 19,307,561 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -39.83 |
| Consensus MFE | -32.52 |
| Energy contribution | -31.88 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
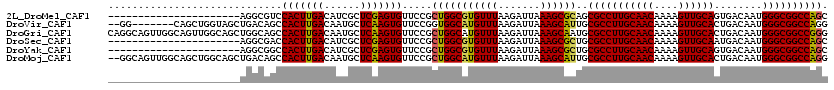
>2L_DroMel_CAF1 19307463 98 + 22407834 ----------------------AGGCGUCCACUUGACAUCGCUCGAGUGUUCCGCUGGCGUGUUUAAGAUUAAAGCGCAGCGCCUUGCAACAAAAGUUGCAGUGACAAUGGGCGGCCAGC ----------------------.((....(((((((......)))))))..))((((((((((((.......))))))..(((((((((((....))))))........))))))))))) ( -38.30) >DroVir_CAF1 873 111 + 1 --GG-------CAGCUGGUAGCUGACAGCCACUUGACAAUGCUCAAGUGUUCCGGUGGCAUGUUUAAGAUUAAAGCAUUGCGCCUUGCAACAAAAGUUGCACUGACAAUGGGCGGCCAGG --((-------(.(((.....((((((((((((.((..((((....)))))).)))))).))))..)).....)))....(((((((((((....))))))........))))))))... ( -38.00) >DroGri_CAF1 1204 120 + 1 CAGGCAGUUGGCAGUUGGCAGCUGGCAGCCACUUGACAAUGCUCAAGUGUUCCGCUGGCAUGUUUAAGAUUAAAGCAAUGCGCCUUGCAACAAAAGUUGCACUGACAAUGGGCGGCCGGG ..(.((((((.(....).)))))).)...(((((((......))))))).....(((((.(((((.......)))))...(((((((((((....))))))........)))))))))). ( -42.90) >DroSec_CAF1 126 98 + 1 ----------------------AGGCGACCACUUGACAUCGCUCGAGUGUUCCGCUGGCGUGUUUAAGAUUAAAGCGCUGCGCCUUGCAACAAAAGUUGCAAUGACAAUGGGCGGCCAGC ----------------------.((.(((.((((((......)))))))))))((((((((((((.......))))))..(((((((((((....))))))........))))))))))) ( -38.20) >DroYak_CAF1 171 98 + 1 ----------------------AGGCGGCCACUUGACAUCGCUCGAGUGUUCCGCUGGCGUGUUUAAGAUUAAAGCGCUGCGCCUUGCAACAAAAGUUGCAGUGACAAUGGGCGGCCAGC ----------------------.(((((.(((((((......)))))))..)))))(((((((((.......))))))..(((((((((((....))))))........))))))))... ( -38.90) >DroMoj_CAF1 17830 118 + 1 --GGCAGUUGGCAGCUGGCAGCUGACAGCCACUUGACAAUGCUCAAGUGUUCCGCUGGCAUGUUUAAGAUUAAAGCAUUGCGCCUUGCAACAAAAGUUGCACUGACAAUGGGCGGCCAGG --((..((((.((((.....)))).))))(((((((......)))))))..)).(((((((((((.......))))))..(((((((((((....))))))........)))))))))). ( -42.70) >consensus ______________________AGGCAGCCACUUGACAACGCUCAAGUGUUCCGCUGGCAUGUUUAAGAUUAAAGCACUGCGCCUUGCAACAAAAGUUGCACUGACAAUGGGCGGCCAGC .............................(((((((......))))))).....(((((((((((.......))))))..(((((((((((....))))))........)))))))))). (-32.52 = -31.88 + -0.64)
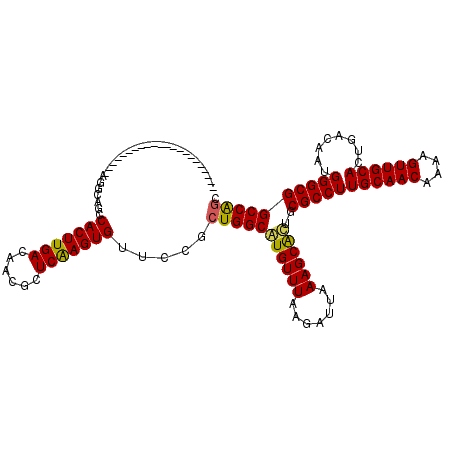

| Location | 19,307,463 – 19,307,561 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -33.79 |
| Consensus MFE | -26.05 |
| Energy contribution | -24.88 |
| Covariance contribution | -1.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
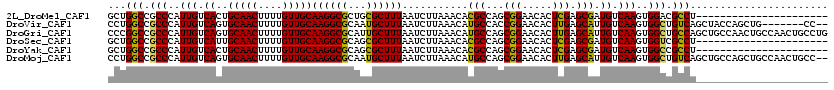
>2L_DroMel_CAF1 19307463 98 - 22407834 GCUGGCCGCCCAUUGUCACUGCAACUUUUGUUGCAAGGCGCUGCGCUUUAAUCUUAAACACGCCAGCGGAACACUCGAGCGAUGUCAAGUGGACGCCU---------------------- ((((((.((.((.((((..((((((....)))))).)))).)).))((((....))))...))))))((..((((.((......)).))))....)).---------------------- ( -31.20) >DroVir_CAF1 873 111 - 1 CCUGGCCGCCCAUUGUCAGUGCAACUUUUGUUGCAAGGCGCAAUGCUUUAAUCUUAAACAUGCCACCGGAACACUUGAGCAUUGUCAAGUGGCUGUCAGCUACCAGCUG-------CC-- ...((((((.....(((..((((((....)))))).)))(((((((((...(((.............)))......)))))))))...))))))..((((.....))))-------..-- ( -32.72) >DroGri_CAF1 1204 120 - 1 CCCGGCCGCCCAUUGUCAGUGCAACUUUUGUUGCAAGGCGCAUUGCUUUAAUCUUAAACAUGCCAGCGGAACACUUGAGCAUUGUCAAGUGGCUGCCAGCUGCCAACUGCCAACUGCCUG ...(((.(....(((.(((((((((....)))))..(((((((.(.((((....))))))))).(((((..(((((((......)))))))....)).)))))).)))).)))).))).. ( -36.90) >DroSec_CAF1 126 98 - 1 GCUGGCCGCCCAUUGUCAUUGCAACUUUUGUUGCAAGGCGCAGCGCUUUAAUCUUAAACACGCCAGCGGAACACUCGAGCGAUGUCAAGUGGUCGCCU---------------------- (((((((((....((((.(((((((....)))))))))))..))).((((....))))...))))))((..((((.((......)).))))....)).---------------------- ( -31.40) >DroYak_CAF1 171 98 - 1 GCUGGCCGCCCAUUGUCACUGCAACUUUUGUUGCAAGGCGCAGCGCUUUAAUCUUAAACACGCCAGCGGAACACUCGAGCGAUGUCAAGUGGCCGCCU---------------------- ((.((((((.(((((((..((((((....))))))...(((.(((.((((....))))..)))..)))........)).)))))....))))))))..---------------------- ( -30.80) >DroMoj_CAF1 17830 118 - 1 CCUGGCCGCCCAUUGUCAGUGCAACUUUUGUUGCAAGGCGCAAUGCUUUAAUCUUAAACAUGCCAGCGGAACACUUGAGCAUUGUCAAGUGGCUGUCAGCUGCCAGCUGCCAACUGCC-- ..((((.((.(((((((..((((((....))))))..).))))))...............((.((((.((.(((((((......)))))))....)).)))).)))).))))......-- ( -39.70) >consensus CCUGGCCGCCCAUUGUCAGUGCAACUUUUGUUGCAAGGCGCAGCGCUUUAAUCUUAAACACGCCAGCGGAACACUCGAGCAAUGUCAAGUGGCCGCCA______________________ ...(((.(((..(((.((..(((((....)))))((((((...))))))...........(((...(((.....))).))).)).)))..))).)))....................... (-26.05 = -24.88 + -1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:18 2006