
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,300,337 – 19,300,490 |
| Length | 153 |
| Max. P | 0.966831 |

| Location | 19,300,337 – 19,300,450 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19300337 113 + 22407834 ACCUCCCUUAAUUUGUAGGGUAACAUUUAUUCGUGAAUUCGAUUUGCUUGCCUUUAAUGUGUCAUCGGAGUUGGUGGGCGGAGAUAAAUGCCACCUGCCGCCCACAACUCACU ................(((((((((.....(((......)))..)).))))))).............((((((.(((((((((...........)).)))))))))))))... ( -31.10) >DroSec_CAF1 41344 113 + 1 ACCUUCCUUAAUUUGUAGGGUAACAUUUAUUCGUGAUUUCGAUUUGCUUGCCUUUAAUGUGUCAUUGGAGUUGCUGGGCGGAGAUAAAUGCCACCUGCCGCCCACAACUCACU ................(((((((((.....(((......)))..)).))))))).............((((((.(((((((((...........)).)))))))))))))... ( -31.10) >DroSim_CAF1 48097 111 + 1 ACCUCCCUUACUUUGUAGGGUAACAUUUAUUCGUGAUUUCGAUUUGCUUGCCUUUAAUGUGUCAU--GAGUCGGUGGGCGGAGAUAAAUGCCACCUGCCGCCCACAACUCACU ........(((.(((.(((((((((.....(((......)))..)).)))))))))).)))...(--((((..((((((((((...........)).)))))))).))))).. ( -32.20) >DroEre_CAF1 46781 112 + 1 ACCUCUCUUGAUAUACAGGGUAUUGUUUAUUCGCGGUUUCGAUUUGCUUACG-CUAUUGUGUCACUGGAGUUGGUGGGCGGAGAUAAAUGCCACCUGCCACCCACAACUCACU ........(((((((.((.(((..((....(((......)))...)).))).-))..)))))))...((((((.((((.((((...........)).)).))))))))))... ( -28.60) >DroYak_CAF1 47372 113 + 1 ACUUUCCCUUAUAAAUAGGGUAUUUUUUAUUCGUGGUUUCGAUUUGCUUACCUUUAUUGUGUCAUUGGAGUUGGUGGGCGGAGAUAAAUGCCUCCUGCCACCCACAACUCACU .........(((((..((((((........(((......)))......))))))..)))))......((((((...(((((((........)))).))).....))))))... ( -27.54) >DroAna_CAF1 70743 111 + 1 UCAUUCACUCGCCUCUAGGGUAUAUUUUAAUCCAAAUUUAGCUUCCUGCUCCUUUUUUGUAUUGCCGG-GCCAGUGGGUGGAGCAGGAAACCACCUGCCGUCCCUGGUUC-UU (((((((((.((((...((((........))))......(((.....)))................))-)).))))))))).(((((......)))))............-.. ( -29.00) >consensus ACCUCCCUUAAUUUGUAGGGUAACAUUUAUUCGUGAUUUCGAUUUGCUUGCCUUUAAUGUGUCAUUGGAGUUGGUGGGCGGAGAUAAAUGCCACCUGCCGCCCACAACUCACU .....(((((.....)))))...............................................((((((.(((((((((...........)).)))))))))))))... (-20.53 = -20.20 + -0.33)
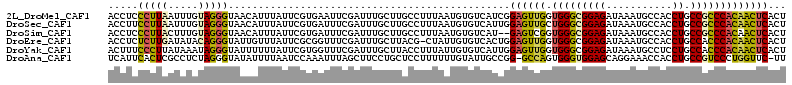

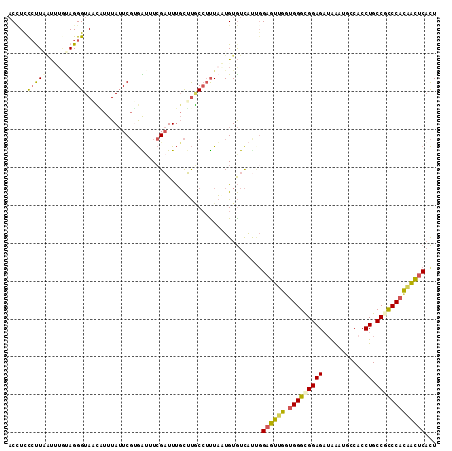
| Location | 19,300,337 – 19,300,450 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -21.91 |
| Energy contribution | -21.88 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19300337 113 - 22407834 AGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCACCAACUCCGAUGACACAUUAAAGGCAAGCAAAUCGAAUUCACGAAUAAAUGUUACCCUACAAAUUAAGGGAGGU .(((((((((((((((.(((((.....)))))))))))))......((((....................)))))))))))............((((........)))).... ( -33.95) >DroSec_CAF1 41344 113 - 1 AGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCAGCAACUCCAAUGACACAUUAAAGGCAAGCAAAUCGAAAUCACGAAUAAAUGUUACCCUACAAAUUAAGGAAGGU .(.(((((((((((((.(((((.....)))))))))))).))))))).............(((..((((..(((......))).....))))..)))................ ( -30.30) >DroSim_CAF1 48097 111 - 1 AGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCACCGACUC--AUGACACAUUAAAGGCAAGCAAAUCGAAAUCACGAAUAAAUGUUACCCUACAAAGUAAGGGAGGU .(((((((((((((((.(((((.....)))))))))))).))))))--))...(((((....(....)...(((......)))...)))))..((((........)))).... ( -35.50) >DroEre_CAF1 46781 112 - 1 AGUGAGUUGUGGGUGGCAGGUGGCAUUUAUCUCCGCCCACCAACUCCAGUGACACAAUAG-CGUAAGCAAAUCGAAACCGCGAAUAAACAAUACCCUGUAUAUCAAGAGAGGU .(.(((((((((((((.(((((.....)))))))))))).)))))))............(-(....))...(((......))).........(((((.(......).)).))) ( -31.90) >DroYak_CAF1 47372 113 - 1 AGUGAGUUGUGGGUGGCAGGAGGCAUUUAUCUCCGCCCACCAACUCCAAUGACACAAUAAAGGUAAGCAAAUCGAAACCACGAAUAAAAAAUACCCUAUUUAUAAGGGAAAGU .(.(((((((((((((..(((........)))))))))).)))))))..............(((...(.....)..)))..............((((.......))))..... ( -28.40) >DroAna_CAF1 70743 111 - 1 AA-GAACCAGGGACGGCAGGUGGUUUCCUGCUCCACCCACUGGC-CCGGCAAUACAAAAAAGGAGCAGGAAGCUAAAUUUGGAUUAAAAUAUACCCUAGAGGCGAGUGAAUGA ..-.....((((........((((((((((((((.((....)).-..(......)......)))))))))))))).((((......))))...))))................ ( -30.00) >consensus AGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCACCAACUCCAAUGACACAAUAAAGGCAAGCAAAUCGAAAUCACGAAUAAAUGAUACCCUACAAAUUAAGGAAGGU ...(((((((((((((.((((((...))))))))))))).)))))).........................(((......)))...........(((........)))..... (-21.91 = -21.88 + -0.02)


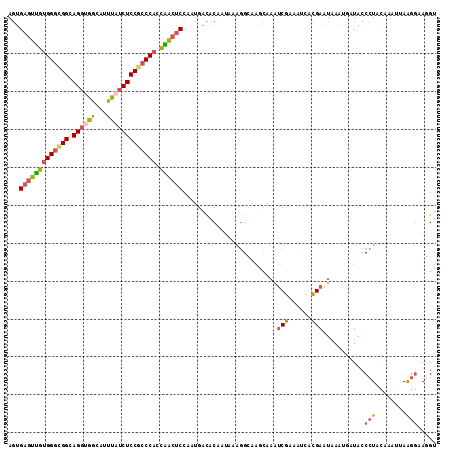
| Location | 19,300,377 – 19,300,490 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -27.55 |
| Energy contribution | -27.58 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19300377 113 - 22407834 AUAAAAAAGGCAUUUGUAAGACAUGAAAGGUACUCAGUCAAGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCACCAACUCCGAUGACACAUUAAAGGCAAGCAAAUC .........((..((((......(((.......)))((((.(.(((((((((((((.(((((.....)))))))))))).)))))))..)))).........))))))..... ( -34.20) >DroSec_CAF1 41384 113 - 1 AUAAAAAAGGCAUUUGUAAGACAUGAAAGGUACUCAGUCAAGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCAGCAACUCCAAUGACACAUUAAAGGCAAGCAAAUC .........((..((((......(((.......)))((((...(((((((((((((.(((((.....)))))))))))).))))))...)))).........))))))..... ( -33.80) >DroSim_CAF1 48137 111 - 1 AUAAAAAAGGCAUUUGUAAGACAUGAAAGGUACUCAGUCAAGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCACCGACUC--AUGACACAUUAAAGGCAAGCAAAUC .........((..((((..(((.(((.......))))))..(((((((((((((((.(((((.....)))))))))))).))))))--))............))))))..... ( -35.80) >DroEre_CAF1 46821 112 - 1 AUAAAAAGGGCAUUUGUAAGACAUGAAAGGUACUCAGUCAAGUGAGUUGUGGGUGGCAGGUGGCAUUUAUCUCCGCCCACCAACUCCAGUGACACAAUAG-CGUAAGCAAAUC .......(((.((((............)))).))).((((...(((((((((((((.(((((.....)))))))))))).))))))...))))......(-(....))..... ( -35.50) >DroYak_CAF1 47412 113 - 1 AUAAAAAAGGCAUUUGUAAGACAUGAAAGGUACUCAGUCAAGUGAGUUGUGGGUGGCAGGAGGCAUUUAUCUCCGCCCACCAACUCCAAUGACACAAUAAAGGUAAGCAAAUC ...........((((((...((.(((.......)))((((...(((((((((((((..(((........)))))))))).))))))...)))).........))..)))))). ( -30.40) >DroAna_CAF1 70783 111 - 1 AUAAAAAAGGCAUUUGUAAGACAUGAAAGGCACUCAGUCAAA-GAACCAGGGACGGCAGGUGGUUUCCUGCUCCACCCACUGGC-CCGGCAAUACAAAAAAGGAGCAGGAAGC ..........(((((((..(((.(((.......))))))...-...(....)...)))))))((((((((((((.((....)).-..(......)......)))))))))))) ( -30.30) >consensus AUAAAAAAGGCAUUUGUAAGACAUGAAAGGUACUCAGUCAAGUGAGUUGUGGGCGGCAGGUGGCAUUUAUCUCCGCCCACCAACUCCAAUGACACAAUAAAGGCAAGCAAAUC ...........((((((..(((.(((.......))))))....(((((((((((((.((((((...))))))))))))).))))))....................)))))). (-27.55 = -27.58 + 0.03)

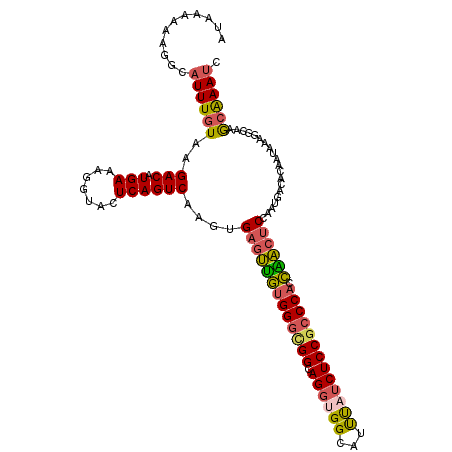

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:12 2006