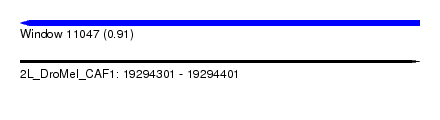
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,294,301 – 19,294,401 |
| Length | 100 |
| Max. P | 0.907635 |

| Location | 19,294,301 – 19,294,401 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 70.37 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -8.58 |
| Energy contribution | -9.33 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
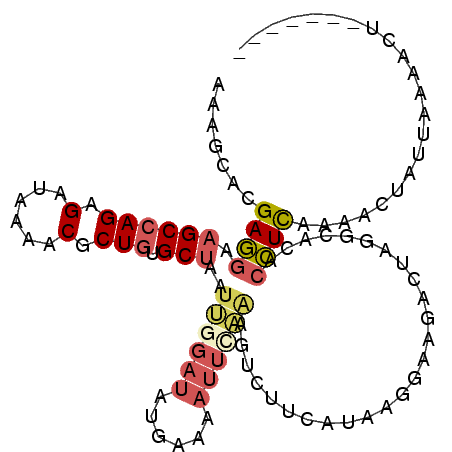
>2L_DroMel_CAF1 19294301 100 - 22407834 AAAGCACGAGGAAGCCAGAGAUAAGUCGCUGUGCUAAUUGGAUAUCAAAAUUCAAAGAGUUCAUAGGGAAGACUAGGCACACCUCACAGCUAUUACAACU------- ..(((..((((..(((..((.....((.(((((((..((((((......))))))..))..))))).))...)).)))...))))...))).........------- ( -23.00) >DroSec_CAF1 35376 100 - 1 AAAGCACGAGGAAGCCAGAGAUAAAUCGCUGUGCUAAUUGGAUAUGAAAAUUCAAAGUCUUCAUAAGGAAGACUAGGCACACCUCACAACCAUUACAACU------- .......((((.(((..((......)))))(((((..((((((......))))))(((((((.....))))))).))))).))))...............------- ( -26.50) >DroSim_CAF1 42210 100 - 1 AAAGCACGAGGAAGCCAGAGACAAGUCGCUGUGCUAAUUGGAUAUGAAAAUUCAAAGUCUUCAUAAGGAAGACUAGGCACACCUCACAACCAUUACAACU------- .......((((.....((.((....)).))(((((..((((((......))))))(((((((.....))))))).))))).))))...............------- ( -26.20) >DroEre_CAF1 41084 87 - 1 GAAGCAAGAGGAAGCCAGAGAUAAAACUCUGUGCUAAUUGGAUGCGAAAAUUGCGAGU-----------AGUUUAGGCACACCUUAA-ACUAUUAA-ACU------- ...((((.....((((((((......))))).)))..(((....)))...)))).(((-----------((((((((.....)))))-))))))..-...------- ( -22.40) >DroYak_CAF1 41331 100 - 1 GAAGCACGAGGAUGCCAGGGAUAAAACUCUGUGCUAAUUGGAUAUAAAAAUUGAUAGUGUUCGUAGGAUGGCUUAGGCACACCUUAAAACUAUUAAAACU------- ..((((((((..(.(....)...)..))).)))))...............((((((((......(((.((((....)).)))))....))))))))....------- ( -21.80) >DroAna_CAF1 62423 90 - 1 CAAGCUCGAAGAAGCUAGAGACAAGACCCUCUGCUAAUAG-----CAAAAUUUUAAGUAUUUGGAAGC----------AAAGUUCAA-AUUGUAGA-AUUCAUAUUU ...(((......(((.((((........)))))))...))-----)..(((((((...(((((((...----------....)))))-))..))))-)))....... ( -14.70) >consensus AAAGCACGAGGAAGCCAGAGAUAAAACGCUGUGCUAAUUGGAUAUGAAAAUUCAAAGUCUUCAUAAGGAAGACUAGGCACACCUCAAAACUAUUAAAACU_______ .......((((.((((((.(......).))).)))..((((((......))))))..........................))))...................... ( -8.58 = -9.33 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:10 2006