| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,293,717 – 19,293,813 |
| Length | 96 |
| Max. P | 0.688296 |

| Location | 19,293,717 – 19,293,813 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -11.37 |
| Energy contribution | -12.65 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
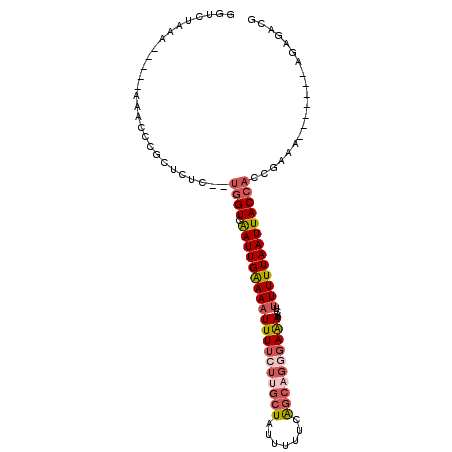
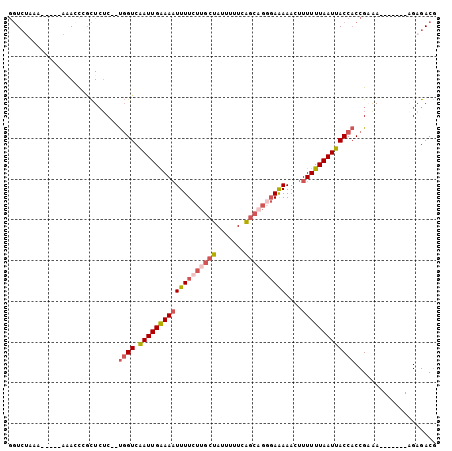
>2L_DroMel_CAF1 19293717 96 - 22407834 GGUCUAAAAAAAAAAACCCGCUCUC--UGGUCAAUUGAAAAUUUUCUUGCUAUUUUUCAGCAGGGAAAAACUUUUUUAAUUACCACCGAAA-------AGAGACG ..................((.((((--((((.(((((((((((((((((((.......))))))))))....)))))))))))).......-------))))))) ( -20.61) >DroSec_CAF1 34766 91 - 1 GGUCUAAA-----AAACCGGCUCGC--UGGUCAAUUGAAAAUUUUCUUGCUAUUUUUCAGCAGGGAAAAACUUUUUUAAUUACCACCGAAA-------AGAGACG .((((...-----....(((.....--((((.(((((((((((((((((((.......))))))))))....))))))))))))))))...-------..)))). ( -24.00) >DroSim_CAF1 41600 91 - 1 GGUCUAAA-----AAACCGGCUCUC--UGGUCAAUUGAAAAUUUUCUUGCUAUUUUUCAGCAGGGAAAAACUUUUUUAAUUACCACCGAAA-------AGAGACG .((((...-----....(((.....--((((.(((((((((((((((((((.......))))))))))....))))))))))))))))...-------..)))). ( -24.00) >DroEre_CAF1 40511 97 - 1 GGUCUAAA-----A-AGUCGCUCUC--UGGUCGAUUGAAAAUUUUCUUGCUAUUUUUCAGCUGUGAAAAACUUUUUUAAUUACCACCGUGACGGUGACGGUGACG ........-----.-.((((((.((--.(((.((((((((((((((..(((.......)))...)))))...))))))))))))((((...)))))).)))))). ( -25.10) >DroYak_CAF1 40785 90 - 1 GGUCUAAA-----AAACUCCCUAUC--UGGUCAAUUGAAAAUUUUCUUGCUAUUUCU-AGCAGUGAAAAACUUUUUUAAUUACCACCGAAA-------AGAGACG .((((...-----............--((((.((((((((((((((((((((....)-))))).)))))...))))))))))))).(....-------.))))). ( -18.90) >DroAna_CAF1 61805 89 - 1 ACUAAAAA-----A-AACAAUUCUCCACUGUCAAUUGGAAUUUUUCUU-CUAUUUUUCGAUU-GCAGAAACUUUUUUAAUUACC-CCGCAA-------CGAGACA ........-----.-......((((..(((.(((((((((........-.....))))))))-))))(((....))).......-......-------.)))).. ( -10.82) >consensus GGUCUAAA_____AAACCCGCUCUC__UGGUCAAUUGAAAAUUUUCUUGCUAUUUUUCAGCAGGGAAAAACUUUUUUAAUUACCACCGAAA_______AGAGACG ...........................((((.(((((((((((((((((((.......))))))))))....))))))))))))).................... (-11.37 = -12.65 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:08 2006