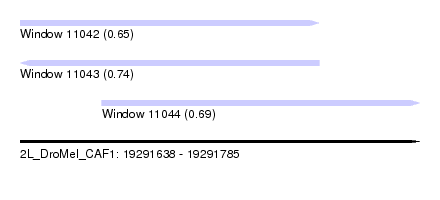
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,291,638 – 19,291,785 |
| Length | 147 |
| Max. P | 0.738692 |

| Location | 19,291,638 – 19,291,748 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -27.67 |
| Energy contribution | -27.84 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
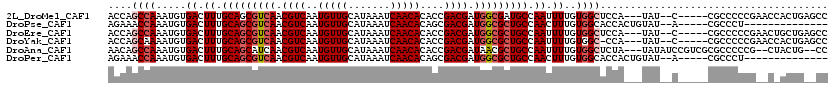
>2L_DroMel_CAF1 19291638 110 + 22407834 GGCUCAGUGGUUCGGGGGCG-----G--AUA---UGGAGCCACAAAAUUGGCAUCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGCUGGU (((((....(((((....))-----)--)).---..))))).........(((.((.((.(((((((((((........)).))))))))).)).)).)))..(((((....)))))... ( -37.20) >DroPse_CAF1 14555 99 + 1 --------------AGGGCG-----U--AUACAGUGGUGCCACAAAGUUGGCAGCGCCAUCGUCGCUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGUUUCU --------------..(((.-----.--.....(((....))).......((((((.((.(((((..((((((.......))))))))))).)).))))))...)))............. ( -31.00) >DroEre_CAF1 38644 110 + 1 GGCUCAGCAGUUCGGGGGCG-----G--AUA---UGGAGCCACAAAAUUGGCAGCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGCUGGU (((((....(((((....))-----)--)).---..))))).........((((((.((.(((((((((((........)).))))))))).)).))))))..(((((....)))))... ( -42.70) >DroYak_CAF1 38741 109 + 1 GGCUCAGUGGUUCGGGGGCG-----G--AUA---UGG-GCCACAAAAUUGGCAGCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUUGCUGGU ((((((...(((((....))-----)--)).---)))-))).((((((((((((((.((.(((((((((((........)).))))))))).)).)))))).....)).))))))..... ( -41.30) >DroAna_CAF1 57055 113 + 1 GG--CAGUAG--CGGGGGCGCGACGGAUAUA---UAGAGCCACAAAAUUGGCAGCGUUAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGAUGCUGCAAAGUCACAUUUGGCUGUU .(--((((.(--((....)))(((((.....---.....)).........(((((((((.(((((((((((........)).))))))))).)))))))))...))).......))))). ( -40.10) >DroPer_CAF1 14929 99 + 1 --------------AGGGCG-----U--AUACAGUGGUGCCACAAAGUUGGCAGCGCCAUCGUCGCUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGUUUCU --------------..(((.-----.--.....(((....))).......((((((.((.(((((..((((((.......))))))))))).)).))))))...)))............. ( -31.00) >consensus GG__CAGU_G__CGGGGGCG_____G__AUA___UGGAGCCACAAAAUUGGCAGCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGCUGGU ................(((...................))).........((((((.((.(((((((((((........)).))))))))).)).))))))..(((((....)))))... (-27.67 = -27.84 + 0.17)

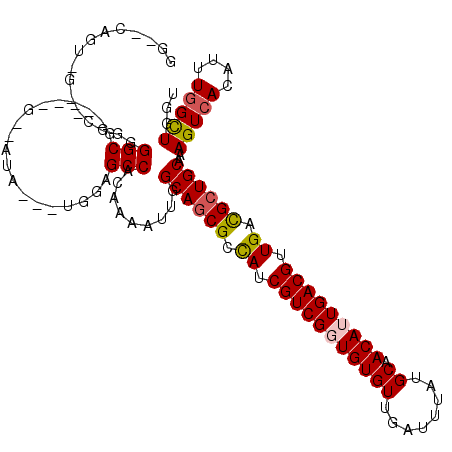
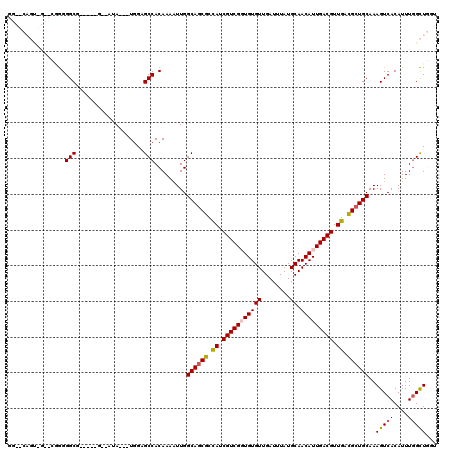
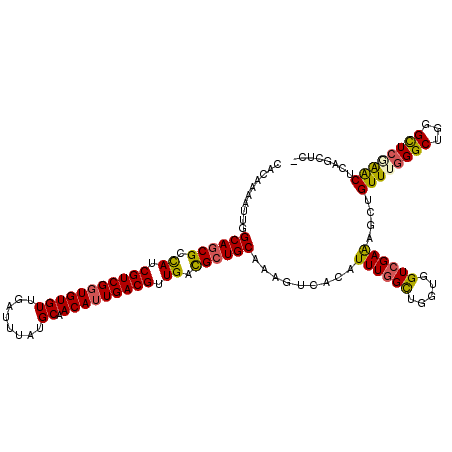
| Location | 19,291,638 – 19,291,748 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19291638 110 - 22407834 ACCAGCCAAAUGUGACUUUGCAGCGUCAACGUCAAUGUUGCAUAAAUCAACACACCGACGAUGGCGAUGCCAAUUUUGUGGCUCCA---UAU--C-----CGCCCCCGAACCACUGAGCC ....((((...(((.....(((.(((((.((((..(((((.......)))))....)))).))))).)))....((((.(((....---...--.-----.)))..)))).))))).)). ( -24.50) >DroPse_CAF1 14555 99 - 1 AGAAACCAAAUGUGACUUUGCAGCGUCAACGUCAAUGUUGCAUAAAUCAACACAGCGACGAUGGCGCUGCCAACUUUGUGGCACCACUGUAU--A-----CGCCCU-------------- ...........(((.....(((((((((.((((..(((((.......)))))....)))).)))))))))..((...(((....))).))..--.-----)))...-------------- ( -29.10) >DroEre_CAF1 38644 110 - 1 ACCAGCCAAAUGUGACUUUGCAGCGUCAACGUCAAUGUUGCAUAAAUCAACACACCGACGAUGGCGCUGCCAAUUUUGUGGCUCCA---UAU--C-----CGCCCCCGAACUGCUGAGCC ..((((.............(((((((((.((((..(((((.......)))))....)))).)))))))))....((((.(((....---...--.-----.)))..))))..)))).... ( -32.90) >DroYak_CAF1 38741 109 - 1 ACCAGCAAAAUGUGACUUUGCAGCGUCAACGUCAAUGUUGCAUAAAUCAACACACCGACGAUGGCGCUGCCAAUUUUGUGGC-CCA---UAU--C-----CGCCCCCGAACCACUGAGCC ....((.....(((.....(((((((((.((((..(((((.......)))))....)))).)))))))))....((((.(((-...---...--.-----.)))..)))).)))...)). ( -30.90) >DroAna_CAF1 57055 113 - 1 AACAGCCAAAUGUGACUUUGCAGCAUCAACGUCAAUGUUGCAUAAAUCAACACACCGACGAUAACGCUGCCAAUUUUGUGGCUCUA---UAUAUCCGUCGCGCCCCCG--CUACUG--CC ...(((....((((....((((((((........))))))))........)))).((((((((.....((((......))))....---..))).))))).......)--))....--.. ( -24.70) >DroPer_CAF1 14929 99 - 1 AGAAACCAAAUGUGACUUUGCAGCGUCAACGUCAAUGUUGCAUAAAUCAACACAGCGACGAUGGCGCUGCCAACUUUGUGGCACCACUGUAU--A-----CGCCCU-------------- ...........(((.....(((((((((.((((..(((((.......)))))....)))).)))))))))..((...(((....))).))..--.-----)))...-------------- ( -29.10) >consensus ACCAGCCAAAUGUGACUUUGCAGCGUCAACGUCAAUGUUGCAUAAAUCAACACACCGACGAUGGCGCUGCCAAUUUUGUGGCUCCA___UAU__C_____CGCCCCCG__C_ACUG__CC ....((((.....((.((.(((((((((.((((..(((((.......)))))....)))).))))))))).)).))..))))...................................... (-23.84 = -24.70 + 0.86)
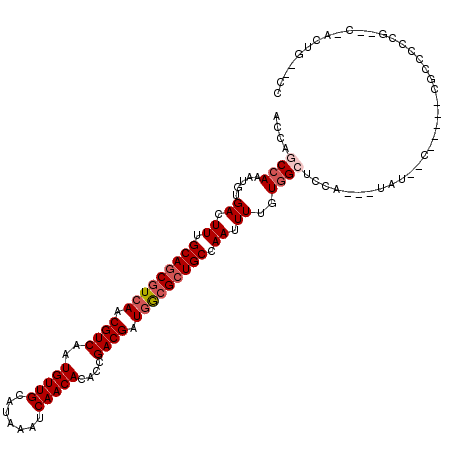

| Location | 19,291,668 – 19,291,785 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.68 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19291668 117 + 22407834 CACAAAAUUGGCAUCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGCUGGUGGUCGAAAGCUGUUUGGGCUGGGCUCGAACUCAGCUC- ..((((...(((.((((((((((((((((((........)).))))))))((.(((........)))))........))))).)))..))).))))(((((((......))))))).- ( -37.70) >DroPse_CAF1 14574 110 + 1 CACAAAGUUGGCAGCGCCAUCGUCGCUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGUUUCUGGUCGAGAGCUGUUGGGGCCUGGUUGGGGC-------- ..........((((((.((.(((((..((((((.......))))))))))).)).))))))...(((.((...(((..(.((.(....)))...)..)))....)).)))-------- ( -35.30) >DroSec_CAF1 32774 117 + 1 CACAAAAUUGGCAGCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGCUGGUGGUCGAAAGCUGUUUGGGCUGGGCUCGAACUCAGCUC- (((.......((((((.((.(((((((((((........)).))))))))).)).))))))..(((((....))))).)))......((((((((((((...))))))))..)))).- ( -43.70) >DroEre_CAF1 38674 116 + 1 CACAAAAUUGGCAGCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGCUGGUGGUCGAGAGCUGUUUGGGCUG-GCUCAAACUCAGCUC- (((.......((((((.((.(((((((((((........)).))))))))).)).))))))..(((((....))))).))).....((((((((((((...-.)))))))..)))))- ( -45.80) >DroYak_CAF1 38770 117 + 1 CACAAAAUUGGCAGCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUUGCUGGUGGUCGAAAGCUGUUUGGGCUGGGCUCAAACUCAGCUC- ..........((((((.((.(((((((((((........)).))))))))).)).)))))).............(((((.((.(....)))((((((((...)))))))))))))..- ( -42.60) >DroAna_CAF1 57088 118 + 1 CACAAAAUUGGCAGCGUUAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGAUGCUGCAAAGUCACAUUUGGCUGUUGGUCGAAAGCUGUUUGGGCCGGGCUCUAACUCCGGUCU ..((((....(((((((((.(((((((((((........)).))))))))).)))))))))..(((((....)))))...((.(....))).))))(((((((.......))))))). ( -43.60) >consensus CACAAAAUUGGCAGCGCCAUCGUCGGUGUGUUGAUUUAUGCAACAUUGACGUUGACGCUGCAAAGUCACAUUUGGCUGGUGGUCGAAAGCUGUUUGGGCUGGGCUCGAACUCAGCUC_ ..........((((((.((.(((((((((((........)).))))))))).)).)))))).........((((((.....))))))....((((((((...))))))))........ (-34.36 = -34.20 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:07 2006