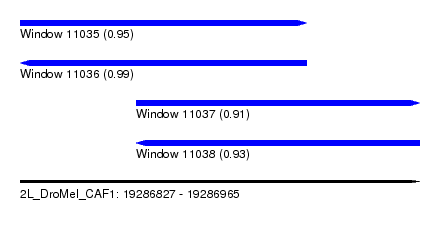
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,286,827 – 19,286,965 |
| Length | 138 |
| Max. P | 0.991344 |

| Location | 19,286,827 – 19,286,926 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -21.54 |
| Energy contribution | -23.40 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19286827 99 + 22407834 AGGUUAAGAAUUUCCCCUUAUCGAUCCCAAUUGCCGCAGGCCAGAGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGA---GGUAAAU----------------GUAGGAUAG ..(((....((((.((((((((((......(((((.(......).)))))(((.((((.((....))))))))))))))))).---)).))))----------------....))).. ( -31.00) >DroSec_CAF1 27939 115 + 1 AGGUUAAGAAUUUCCCCUUAUCGAUCCCAAUUGCCGCGGGCCAGGAGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGA---GGGAAAUGCGCACAGUAGGCAAAGUGGGAUAG .........((((((((((((((((((.....(((...)))..)))....(((.((((.((....))))))))))))))))).---))))))).(.(((..........))).).... ( -35.90) >DroSim_CAF1 34747 115 + 1 AGGUUAAGAAUUUCCCCUUAUCGAUCCCAAUUGCCGUGGGCCACAGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGA---GGGAAAUGCGCACAGUAGGCAAUGUGGGAUAG .........(((((((((((((((..(((.((.(((((((((((.......))).)))))))))).))).....)))))))).---))))))).(.((((........)))).).... ( -43.40) >DroEre_CAF1 33867 109 + 1 AGGUUAAGAAUUUCCCCUUAUCGAUCCCAAUUGCCGCGGG-CAGAGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGA---GGAAAAUGCGA-----AGGCACAGUAGGAUGG ..........(((((.((((((((......(((((.(...-..).)))))(((.((((.((....))))))))))))))))).---))))).(((..-----..)))........... ( -37.20) >DroYak_CAF1 33876 110 + 1 AGUUUAAGAAUUUCCCCUUAUCGAUUCCAAUUGCCGCGGGGAAGAGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGA---GGGAAAUGCGA-----AGGCAAAGUGGGAUGG .........(((((((((((((((......(((((.(......).)))))(((.((((.((....))))))))))))))))).---)))))))((..-----..))............ ( -40.80) >DroPer_CAF1 9956 91 + 1 UU-------GUUGCCUCUUAUCGAUCCCAGGUGCCGCAGGGCAAUGGCAAGGUGAGCCUAUGGAAUU-UCGGGGCCGAUAAGCAUUGGGGAAAGAGA-----AG-------------- ..-------.((.((((((((((..(((...((((....))))..(((.......))).........-..)))..)))))))....))).)).....-----..-------------- ( -28.30) >consensus AGGUUAAGAAUUUCCCCUUAUCGAUCCCAAUUGCCGCGGGCCAGAGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGA___GGGAAAUGCGA_____AGGCAAAGUGGGAUAG .........(((((((((((((((......(((((..........)))))(((.((((.........)))))))))))))))....)))))))......................... (-21.54 = -23.40 + 1.86)

| Location | 19,286,827 – 19,286,926 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -21.05 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19286827 99 - 22407834 CUAUCCUAC----------------AUUUACC---UCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCUCUGGCCUGCGGCAAUUGGGAUCGAUAAGGGGAAAUUCUUAACCU .........----------------.....((---(((((((((..(.(((....(((.((((.......)))).))).....))).....)..)))))))))))............. ( -30.10) >DroSec_CAF1 27939 115 - 1 CUAUCCCACUUUGCCUACUGUGCGCAUUUCCC---UCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCUCCUGGCCCGCGGCAAUUGGGAUCGAUAAGGGGAAAUUCUUAACCU ............(((....).))(.(((((((---.((((((((((.((((.........))))..))....((((.(((...)))....))))))))))))))))))).)....... ( -33.10) >DroSim_CAF1 34747 115 - 1 CUAUCCCACAUUGCCUACUGUGCGCAUUUCCC---UCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCUGUGGCCCACGGCAAUUGGGAUCGAUAAGGGGAAAUUCUUAACCU ......((((........)))).(.(((((((---.((((((((...........((((((((.......))))))))((((.......)))).))))))))))))))).)....... ( -39.70) >DroEre_CAF1 33867 109 - 1 CCAUCCUACUGUGCCU-----UCGCAUUUUCC---UCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCUCUG-CCCGCGGCAAUUGGGAUCGAUAAGGGGAAAUUCUUAACCU ..........((((..-----..))))(((((---(((((((((((...))..(((((.((((.......)))).((-((...))))..))))))))))))))))))).......... ( -33.90) >DroYak_CAF1 33876 110 - 1 CCAUCCCACUUUGCCU-----UCGCAUUUCCC---UCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCUCUUCCCCGCGGCAAUUGGAAUCGAUAAGGGGAAAUUCUUAAACU ...........(((..-----..)))((((((---.((((((((.(.((((.........))))).(((((((.(......).)))))..))..)))))))))))))).......... ( -31.90) >DroPer_CAF1 9956 91 - 1 --------------CU-----UCUCUUUCCCCAAUGCUUAUCGGCCCCGA-AAUUCCAUAGGCUCACCUUGCCAUUGCCCUGCGGCACCUGGGAUCGAUAAGAGGCAAC-------AA --------------..-----...............(((((((..((((.-.........(((.......)))..((((....))))..))))..))))))).(....)-------.. ( -27.10) >consensus CUAUCCCACUUUGCCU_____UCGCAUUUCCC___UCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCUCUGGCCCGCGGCAAUUGGGAUCGAUAAGGGGAAAUUCUUAACCU .........................(((((((....((((((((....(((.........)))...((.((((.(......).))))...))..)))))))))))))))......... (-21.05 = -21.72 + 0.67)



| Location | 19,286,867 – 19,286,965 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.48 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -6.53 |
| Energy contribution | -7.08 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.19 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19286867 98 + 22407834 CCAG--AGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGAGGUAAAU----------------GUAGGAUAGGGUGGGGGAA-AAUUAAGAGUCCGCCCCCCCCCACAAACG (((.--((((.......)))).))).......((((((.....))))))...----------------........(((.(((((..-.((.....))...))))).)))....... ( -33.50) >DroSec_CAF1 27979 111 + 1 CCAG--GAGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGAGGGAAAUGCGCACAGUAGGCAAAGUGGGAUAGGGUGGGGGAAAAAUUAAGAGUUCC----CCCGCCCAGGCG (((.--..(((.....((((.((....)))))).((((.....))))....)))((.......))....)))....(((((((((((..........))))----)))))))..... ( -41.60) >DroSim_CAF1 34787 110 + 1 CCAC--AGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGAGGGAAAUGCGCACAGUAGGCAAUGUGGGAUAGGGUGGGGGAAAAAUUAAGAGUUCC-----CCGCCCAGUCG ((((--(.((...(((((((.((....)))))).((((.....))))........))).....))..)))))(((.((((((((((...........))))-----)))))).))). ( -45.10) >DroEre_CAF1 33907 101 + 1 -CAG--AGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGAGGAAAAUGCGA-----AGGCACAGUAGGAUGGGGUUGGGGAAAAAUUAGGAGGUCC-----CC---UCGUCG -((.--((((.......)))).))......((((((((.....))))....(((..-----..)))))))..(((((....(((((............)))-----))---))))). ( -29.90) >DroYak_CAF1 33916 101 + 1 GAAG--AGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGAGGGAAAUGCGA-----AGGCAAAGUGGGAUGGGGU-GGGGAAAAAUUAAGAGUUCU-----CU---CCAUCG ...(--(((((......(((.((....))))))))))).............(((..-----..)))......(((((((.-(((.............))).-----))---))))). ( -28.32) >DroAna_CAF1 52839 81 + 1 GAAUGCCGCCAAGGUGAGCCUAUA-AAAUUUCAGACUCGAUAAGCGG-------------------------CAUGGCGUGGGGCAA--A-UGAGAAUUUU----CUCG---CGUCG ..(((((((.....((((.((...-.......)).))))....))))-------------------------)))(((((((((.((--(-(....)))))----))))---)))). ( -28.40) >consensus CCAG__AGGCAAGGUGAGCCUAUGGAAAUGGCUGCCUCGAUAAGAGGGAAAUGCG______AGGCAAAGUGGGAUAGGGUGGGGGAAAAAUUAAGAGUUCC_____CCG__CAGUCG ............((..((((.........)))).((((.....))))...........................................................))......... ( -6.53 = -7.08 + 0.56)


| Location | 19,286,867 – 19,286,965 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.48 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -6.30 |
| Energy contribution | -6.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.23 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19286867 98 - 22407834 CGUUUGUGGGGGGGGGCGGACUCUUAAUU-UUCCCCCACCCUAUCCUAC----------------AUUUACCUCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCU--CUGG .((..(((((((((((..((.......))-..))))).))))))...))----------------.....((((.....)))).........(((.((((.......))))--.))) ( -33.60) >DroSec_CAF1 27979 111 - 1 CGCCUGGGCGGG----GGAACUCUUAAUUUUUCCCCCACCCUAUCCCACUUUGCCUACUGUGCGCAUUUCCCUCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCUC--CUGG .((..(((.(((----((((..........))))))).)))...........(((((..(((.((.....((((.....))))..))))).....))))).......))..--.... ( -32.70) >DroSim_CAF1 34787 110 - 1 CGACUGGGCGG-----GGAACUCUUAAUUUUUCCCCCACCCUAUCCCACAUUGCCUACUGUGCGCAUUUCCCUCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCU--GUGG .((..(((.((-----((...............)))).)))..))......((((......).)))....((((.....)))).........((((((((.......))))--)))) ( -33.06) >DroEre_CAF1 33907 101 - 1 CGACGA---GG-----GGACCUCCUAAUUUUUCCCCAACCCCAUCCUACUGUGCCU-----UCGCAUUUUCCUCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCU--CUG- .((.((---((-----(((............)))))..............((((..-----..))))..)).)).....(((((((((.........)))......)))))--)..- ( -22.50) >DroYak_CAF1 33916 101 - 1 CGAUGG---AG-----AGAACUCUUAAUUUUUCCCC-ACCCCAUCCCACUUUGCCU-----UCGCAUUUCCCUCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCU--CUUC .(((((---((-----(((........)))))))..-..............(((..-----..)))..........)))(((((((((.........)))......)))))--)... ( -19.10) >DroAna_CAF1 52839 81 - 1 CGACG---CGAG----AAAAUUCUCA-U--UUGCCCCACGCCAUG-------------------------CCGCUUAUCGAGUCUGAAAUUU-UAUAGGCUCACCUUGGCGGCAUUC .....---.(((----(....)))).-.--............(((-------------------------(((((....(((((((......-..))))))).....)))))))).. ( -24.20) >consensus CGACGG__CGG_____GGAACUCUUAAUUUUUCCCCCACCCCAUCCCACUUUGCCU______CGCAUUUCCCUCUUAUCGAGGCAGCCAUUUCCAUAGGCUCACCUUGCCU__CUGG ..............................................................................(((((.((((.........))))..)))))......... ( -6.30 = -6.97 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:01 2006