| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,281,884 – 19,281,991 |
| Length | 107 |
| Max. P | 0.865305 |

| Location | 19,281,884 – 19,281,991 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -18.60 |
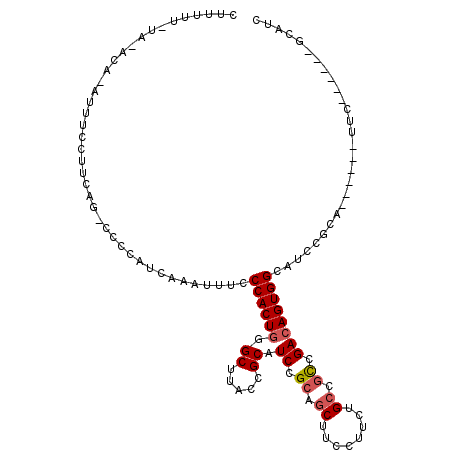
| Consensus MFE | -13.14 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19281884 107 - 22407834 CUUUUUUUUAACA-AUUUCCUUCAG-CUCCAUCAAAUUUCCCACUGGGCUUACCGCAUCCGCAGCUUCCUUCUGCCGCCGACAGUGGGAUCCGCAUUCGCAUUC------GCAUC .............-..........(-(...........((((((((.((.....)).((.((.((........)).)).))))))))))...((....))....------))... ( -24.50) >DroSec_CAF1 22921 99 - 1 CUUUUU-UA-ACA-AUUUCCUUCAG-CCCCAUCAAAUUUCCCACUGGGCUUACCGCAUCCGCAGCUUCCAUCUGCCGCCGACAGUGGCAUCCGCA------CUC------GCAUC ......-..-...-.........((-(((................)))))..((((.((.((.((........)).)).))..)))).....((.------...------))... ( -17.09) >DroSim_CAF1 29827 100 - 1 CUUUUU-UAAACA-AUUUCCUUCAG-CCCCAUCAAAUUUCCCACUGGGCUUACCGCAUCCGCAGCUUCCUUCUGCCGCCGACAGUGGCAUCCGCA------UUC------GCAUC ......-......-.........((-(((................)))))..((((.((.((.((........)).)).))..)))).....((.------...------))... ( -17.09) >DroEre_CAF1 29218 104 - 1 UUUUUU-UA-ACA-AUUUCCUUCAG-CUCCAUCAAAUUUCCCACUGGGCUUACCGCAUCCGCAGCUUCCUUCUGCCGCCGACAGUGGCAUCCGCAUCCGCACUCGCAU------- ......-..-...-..........(-(.............((((((.((.....)).((.((.((........)).)).)))))))).....((....))....))..------- ( -19.20) >DroYak_CAF1 29160 112 - 1 CUUCUU-UA-ACAAAUUUCCUUCAA-CUCCAUCAAACUUCCCACUGGGCUUACCGCAUCCGCAUCUUCCUUCUGCCGCCGACAGUGGCAUCAGCAUCCGCAUUCGCAUCCGCAUC ......-..-...............-...................(((((....((....))..........((((((.....))))))..)))....((....))..))..... ( -15.20) >DroAna_CAF1 45320 86 - 1 ACUUUC-CA-GCC-AUUUCCUACGGUCCCCUUCAAAUUUCCCACUCGGCUUGUGGCAUCCGCUGCCGUCUUCUGCUCUCGACAGUGGCA-------------------------- ......-..-(((-(((.............................(((..((((...)))).)))(((..........))))))))).-------------------------- ( -18.50) >consensus CUUUUU_UA_ACA_AUUUCCUUCAG_CCCCAUCAAAUUUCCCACUGGGCUUACCGCAUCCGCAGCUUCCUUCUGCCGCCGACAGUGGCAUCCGCA______UUC______GCAUC ........................................((((((.((.....)).((.((.((........)).)).))))))))............................ (-13.14 = -13.50 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:54 2006