
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,280,763 – 19,280,871 |
| Length | 108 |
| Max. P | 0.552090 |

| Location | 19,280,763 – 19,280,871 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.35 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19280763 108 - 22407834 AU-GCCGUAAUUAAUGGCCCAUCGC-----------CGGCAUCGCGUUAAUGGACAGAUGCUGAGCCCGCGACCCUUGACCCUCCGCACAAUGCGAUAAGGUCUUCAUUUGGCAUUCUCA ((-((((.....(((((....((((-----------.(((.(((((((........))))).))))).)))).....((((...(((.....)))....)))).)))))))))))..... ( -32.00) >DroPse_CAF1 1353 100 - 1 GU-GCCGUAAUUAAUGGCCCAUCGCAG--------UCGGCAUCGUUUUAAUGGACAGAUGCUGGGCCU-----------CUCUCUGCAUAAAGCGAUAAGGUCUUCAUUUGGCAUUCUCA ((-((((.(((.((.((((.(((((..--------((((((((((((....)))).))))))))((..-----------......)).....)))))..)))))).)))))))))..... ( -31.60) >DroEre_CAF1 28172 107 - 1 AU-GCCGUAAUUAAUGGCCCAUCGC-----------CGGCAUCGCUUUAAUGGACAGAUGCUGAGCCCGCGACCCUUGACCCU-CGCACAAUGCGAUAAGGUCUUCAUUUGGCAUUCUCA ((-((((.(((.((.((((.(((((-----------(((((((.((.....))...))))))).....((((..........)-))).....)))))..)))))).)))))))))..... ( -32.20) >DroYak_CAF1 28040 109 - 1 UGGCCCGUAAUUAAUGGCCCAUCGC-----------CGGCAUCGCUUUAAUGGACAGAUGCUGAGCCCGCGACCCUUGACCCUCCGCACAAUGCGAUAAGGUCUUCAUUUGGCAUUCUCA (((.((((.....)))).)))..((-----------(((((((.((.....))...))))))).))..((.(.....((((...(((.....)))....))))......).))....... ( -29.90) >DroAna_CAF1 44201 114 - 1 GU-GCCGUAAUUAAUGGCCCAUCUCGGGUCCCGGCUCGGCAUCGUUUUAAUGGACAGAUGCCGGGCGG-----ACUGGAAGCGCCGCAUAAAGCGAUAAGGUCUUCAUUUGGCAUUCUCA ((-((((.(((.((.((((....((.(((((..((((((((((((((....)))).))))))))))))-----))).)).....(((.....)))....)))))).)))))))))..... ( -45.50) >DroPer_CAF1 1715 100 - 1 GU-GCCGUAAUUAAUGGCCCAUCGCAG--------UCGGCAUCGUUUUAAUGGACAGAUGCUGGGCCU-----------CUCUCUGCAUAAAGCGAUAAGGUCUUCAUUUGGCAUUCUCA ((-((((.(((.((.((((.(((((..--------((((((((((((....)))).))))))))((..-----------......)).....)))))..)))))).)))))))))..... ( -31.60) >consensus GU_GCCGUAAUUAAUGGCCCAUCGC___________CGGCAUCGCUUUAAUGGACAGAUGCUGAGCCC_____CCUUGACCCUCCGCACAAAGCGAUAAGGUCUUCAUUUGGCAUUCUCA ...((((.(((.((.((((.(((((...........(((((((.............))))))).((...................)).....)))))..)))))).)))))))....... (-23.15 = -23.35 + 0.20)

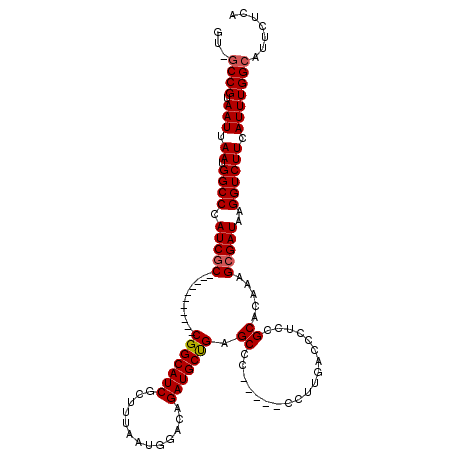
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:53 2006