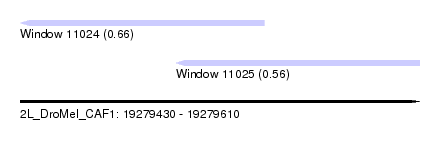
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,279,430 – 19,279,610 |
| Length | 180 |
| Max. P | 0.661300 |

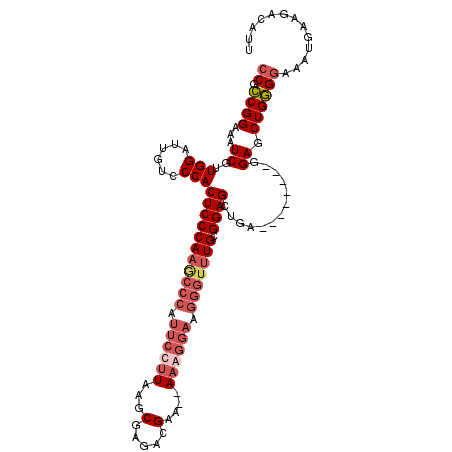
| Location | 19,279,430 – 19,279,540 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -22.66 |
| Energy contribution | -25.23 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19279430 110 - 22407834 CCGCCGGAAAUCGUUGGAUUGUCCCACUCCCAAACCCAUUCCUUAAGCGAGACGAA--AAAGGAAGGGUUUGCGGAGCUGAGGAACUGAGGAGCUGGGGAAAUGAAGACAUU ((.((((...((.(..(....((((((((((((((((.((((((...((...))..--.))))))))))))).)))).)).))).)..).)).))))))............. ( -42.20) >DroSec_CAF1 20536 104 - 1 CCGCCGGAAAUCGUUGGAUUGUCCCACUCCCAAGCCCAUUCCUUAAGCGAGACGAAUGAAGGGAAGGGUUUGCGGAGCUGA--------GGAGCUGGGAAAAUGAAGACAUU ...((((.((((....)))).((((((((((((((((.((((((...((...))....)))))).))))))).)))).)).--------))).))))............... ( -35.60) >DroSim_CAF1 27489 104 - 1 CCGCCGGAAAUCGUUGGAUUGUCCCACUCCCAAGCCCAUUCCUUAAGCGAGACGAAUGAAGGGAAGGGUUUGCGGAGCUGA--------GGAGCUGGGGAAAUGAAGACAUU ((.((((.((((....)))).((((((((((((((((.((((((...((...))....)))))).))))))).)))).)).--------))).))))))............. ( -38.00) >DroEre_CAF1 26979 90 - 1 CCGGCGGGAAUCGUUGGAUUGUCCCACUCCCAC-----------UAUCCAGACGAA---AAGGAAGGGUUUGUGGAGCCGU--------GGAGCUGUGGAAAUGAAGACAUU ((((((.....))))))..((((((((..((((-----------.((((...(...---..)...))))..))))(((...--------...))))))).......)))).. ( -24.01) >consensus CCGCCGGAAAUCGUUGGAUUGUCCCACUCCCAAGCCCAUUCCUUAAGCGAGACGAA__AAAGGAAGGGUUUGCGGAGCUGA________GGAGCUGGGGAAAUGAAGACAUU ((.((((...((..(((......)))(((((((((((.((((((...(.....)....)))))).))))))).)))).............)).))))))............. (-22.66 = -25.23 + 2.56)

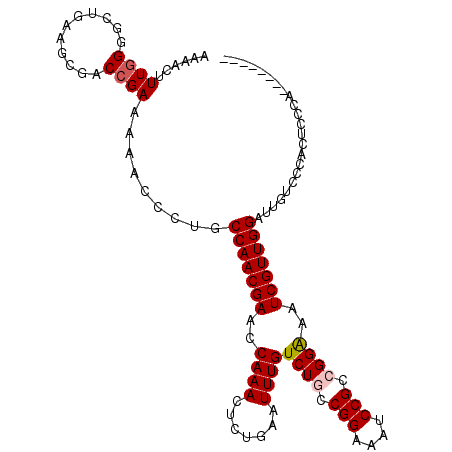
| Location | 19,279,500 – 19,279,610 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19279500 110 - 22407834 AAAACUUUGGGGCUGAAGCGACCGAAAAACCCUGCCAACGAACCAAACUCUGAAUUUGUCUGCCGGAAAUCCGCCGGAAAUCGUUGGAUUGUCCCACUCCCAAACCCAUU .....((((((..((..((((.............(((((((..((((.......))))((((.(((....))).))))..))))))).))))..))..))))))...... ( -27.44) >DroPse_CAF1 32 94 - 1 GAAA-CUUGGAGCGGAAAAAACCGAAAAACCAAGCCAACGAACCAAACUCAGAAUUUGUCUGCCGGAAGUCCGCAGGAAAUCGUUGGAUUCUCAU--------------- (((.-(((((..(((......))).....)))))(((((((..((((.......))))(((((.((....)))))))...))))))).)))....--------------- ( -26.20) >DroSim_CAF1 27553 109 - 1 AAAACUUUGGGGCUGAAGCGACCGAA-AACCCUGCCAACGAACCAAACUCUGAAUUUGUCUGCCGGAAAUCCGCCGGAAAUCGUUGGAUUGUCCCACUCCCAAGCCCAUU .....((((((..((..((((.....-.......(((((((..((((.......))))((((.(((....))).))))..))))))).))))..))..))))))...... ( -27.22) >DroEre_CAF1 27036 103 - 1 AAAACUUUGGGGCUGCAGCGACCGAAAAACCCUGCCAACGAGCCAAACUCUGAAUUUGUCUGCCGGAAAUCCGGCGGGAAUCGUUGGAUUGUCCCACUCCCAC------- .......((((((.((((.(..........)))))((((((..((((.......))))((((((((....))))))))..))))))....)))))).......------- ( -33.40) >DroYak_CAF1 26765 103 - 1 AAAACUUUUGGGCUGAACCGACCGAAAAACCCUGCCAACGAACCAAACUCUGAAUUUGUCUGCCGGAAAUCCGCCGGAAAUCGUUGGAUUGUCCCACUCCCAC------- .....((((((..........)))))).......(((((((..((((.......))))((((.(((....))).))))..)))))))................------- ( -21.90) >DroPer_CAF1 400 94 - 1 GAAA-CUUGGAGCGGAAAAAACCGAAAAACCAAGCCAACGAACCAAACUCAGAAUUUGUCUGCCGGAAGUCCGCAGGAAAUCGUUGGAUUCUCAU--------------- (((.-(((((..(((......))).....)))))(((((((..((((.......))))(((((.((....)))))))...))))))).)))....--------------- ( -26.20) >consensus AAAACUUUGGGGCUGAAGCGACCGAAAAACCCUGCCAACGAACCAAACUCUGAAUUUGUCUGCCGGAAAUCCGCCGGAAAUCGUUGGAUUGUCCCACUCCCA________ ......((((...........)))).........(((((((..((((.......))))((((.(((....))).))))..)))))))....................... (-17.64 = -18.00 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:49 2006