
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,277,736 – 19,277,905 |
| Length | 169 |
| Max. P | 0.950264 |

| Location | 19,277,736 – 19,277,832 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19277736 96 + 22407834 UUUGAUCAAAAAAUGUCAAUUAAAUGGCACUUUAAUAUUAAAUUCAUAAACGACAAC-------------------GAG----UGUGUGUGUGGCAUACAUUGGCACAUAAAUUAUUUA ((((((...(((.(((((......))))).)))...))))))...............-------------------(((----((..((((((.((.....)).))))))...))))). ( -16.50) >DroSec_CAF1 18890 94 + 1 UUUGAUCAAAAAAUGUCAAUUAAAUGGCACUUUAAUAUUAAAUUCAUAAACGACAGC-------------------GAG----UGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUA ((((((...(((.(((((......))))).)))...))))))...............-------------------.((----(.(--((((((((.....))))))))).)))..... ( -19.10) >DroSim_CAF1 25830 94 + 1 UUUGAUCAAAAAAUGUCAAUUAAAUGGCACUUUAAUAUUAAAUUCAUAAACGACAGC-------------------GAG----UGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUA ((((((...(((.(((((......))))).)))...))))))...............-------------------.((----(.(--((((((((.....))))))))).)))..... ( -19.10) >DroEre_CAF1 25347 115 + 1 UUUGAUCAA-AAAUGUCAAUUAAAU-GCACUUUAAUAUUAAAUUCAUAAACGACAGCACGACGACUAAGUGCGUGUGUGUCUGUGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUA .(((((...-....)))))......-((((((((....)))).........(((((((((((......)).))))).)))).))))--((((((((.....)))))))).......... ( -27.30) >DroYak_CAF1 25026 112 + 1 UUUGAUCAAAAAAUGUCAAUUAAAUGGCACUUUAAUAUUAAAUUCAUAAACGA-AACACGACGACUAUGUGCGUGCGUG----UGU--GUCUGGCAUACAUUGCCACAUAAAUUAUUUA .(((((........)))))......(((((............(((......))-)((((((((.(.....)))).))))----)))--)))(((((.....)))))............. ( -22.70) >consensus UUUGAUCAAAAAAUGUCAAUUAAAUGGCACUUUAAUAUUAAAUUCAUAAACGACAGC___________________GAG____UGU__GUGUGGCAUACAUUGCCACAUAAAUUAUUUA ((((((...(((.(((((......))))).)))...))))))..............................................((((((((.....)))))))).......... (-13.50 = -14.30 + 0.80)



| Location | 19,277,776 – 19,277,872 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -16.69 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19277776 96 + 22407834 AAUUCAUAAACGACAAC--------------------GAG----UGUGUGUGUGGCAUACAUUGGCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCU .................--------------------(((----((..((((((.((.....)).))))))...)))))..........(((((...(((((....)))))...))))). ( -21.70) >DroSec_CAF1 18930 94 + 1 AAUUCAUAAACGACAGC--------------------GAG----UGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCU .................--------------------.((----(.(--((((((((.....))))))))).)))..............(((((...(((((....)))))...))))). ( -24.30) >DroSim_CAF1 25870 94 + 1 AAUUCAUAAACGACAGC--------------------GAG----UGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCU .................--------------------.((----(.(--((((((((.....))))))))).)))..............(((((...(((((....)))))...))))). ( -24.30) >DroEre_CAF1 25385 117 + 1 AAUUCAUAAACGACAGCACGACGACUAAGUGC-GUGUGUGUCUGUGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCAU ....((((.(((...(((((((......)).)-)))).))).))))(--((((((((.....)))))))))..................((..(...(((((....)))))..)..)).. ( -33.50) >DroYak_CAF1 25066 112 + 1 AAUUCAUAAACGA-AACACGACGACUAUGUGC-GUGCGUG----UGU--GUCUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAAUGGGGACGAGCAGAAAGUUUUGCAAUUUCCCU ...........((-.((((((((.(.....))-)).))))----)..--.))(((((.....)))))......................(((((...(((((....)))))...))))). ( -26.60) >DroAna_CAF1 41158 110 + 1 AAUUCAUAAACGGAAACCC----AGUCUGGGCUGAGGGAG----UGA--GUGUGGCAUACAUUGCCGCAUAAAUUAUUUAACAUAAAAUGGGGCCGAGCACAAAGUUUUGCAAUUUGCAU ..........(((...(((----(......(.((((..((----(..--((((((((.....))))))))..))).)))).)......)))).))).(((........)))......... ( -28.20) >consensus AAUUCAUAAACGACAAC____________________GAG____UGU__GUGUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCU .................................................((((((((.....))))))))..................((....)).(((((....)))))......... (-16.69 = -16.83 + 0.14)


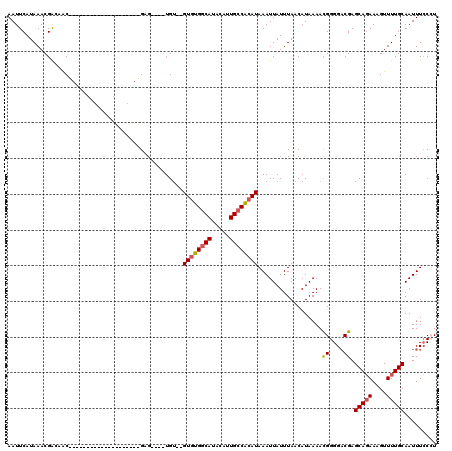
| Location | 19,277,796 – 19,277,905 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
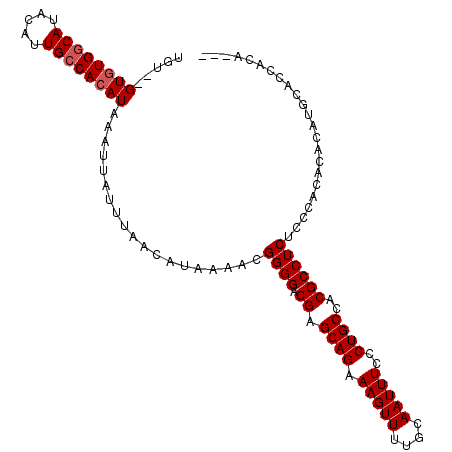
>2L_DroMel_CAF1 19277796 109 + 22407834 UGUGUGUGUGGCAUACAUUGGCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCUGCCACGCCUCUCUCACACACAUACACCAUACAC ((((((((((.........(((.......((((.....))))..((((((...(((((....)))))....))))))....))).......))))))))))........ ( -29.79) >DroSec_CAF1 18950 104 + 1 UGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCUGCCACGCCUCUCCCACACACAUGCACCACA--- (((--((((((((.....))))......................((((((...(((((....)))))....)))))).............))))))).........--- ( -27.00) >DroSim_CAF1 25890 104 + 1 UGU--GUGUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCUGCCACGCCUCUCCCACACACAUGCACCACA--- (((--((((((((.....))))......................((((((...(((((....)))))....)))))).............))))))).........--- ( -27.00) >DroYak_CAF1 25104 101 + 1 UGU--GUCUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAAUGGGGACGAGCAGAAAGUUUUGCAAUUUCCCUGCCCCGCCUCUCACGCCUAC------CACACAC (((--((.(((((.....)))))......................((((.((.((((.(((((....)))))..))))..))))))..........------.))))). ( -22.20) >consensus UGU__GUGUGGCAUACAUUGCCACAUAAAUUAUUUAACAUAAAACGGGGACGAGCAGAAAGUUUUGCAAUUUCCCUGCCACGCCUCUCCCACACACAUGCACCACA___ .....((((((((.....))))))))...................((((.((.((((.(((((....)))))..))))..))))))....................... (-20.83 = -21.32 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:47 2006