
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,272,745 – 19,272,915 |
| Length | 170 |
| Max. P | 0.996972 |

| Location | 19,272,745 – 19,272,850 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.05 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
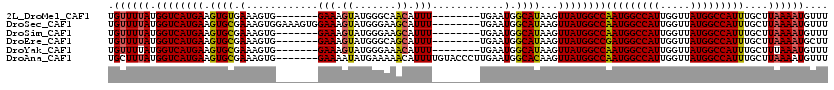
>2L_DroMel_CAF1 19272745 105 + 22407834 UGUUUUAUGGUCAUGAAGUGUGAAAGUG-------GAAAGUAUGGGCAACAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUU .(((((((((((((((..((((....(.-------((((.....(....).)))--------).)....))))..)))))))))((((((((.....)))))))).....)))))).... ( -32.20) >DroSec_CAF1 13978 112 + 1 UGUUUUAUGGUCAUGAAGUGCGAAAGUGGAAAGUGGAAAGUAUGGGAAGCAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUU .....(((..((((.....((....)).....))))...)))....((((((((--------((((((((....))))).((.(((((((((.....))))))))).))))))))))))) ( -31.20) >DroSim_CAF1 20858 105 + 1 UGUUUUAUGGUCAUGAAGUGCGAAAGUG-------GAAAGUAUGGGAAGCAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUU ..........(((((....((....)).-------.....))))).((((((((--------((((((((....))))).((.(((((((((.....))))))))).))))))))))))) ( -29.90) >DroEre_CAF1 20730 105 + 1 UGUUUUAUGGUCAUGAAGUGCGAAAGUG-------GAAAGUAUGGGCAGCAUUU--------UGAAUGGCAUAAGUUAUGGCCGAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGCUU .......((.(((((....((....)).-------.....))))).))((((((--------((((((((....))))).((.(((((((((.....))))))))).))))))))))).. ( -34.40) >DroYak_CAF1 20021 105 + 1 UGUUUUAUGGUCAUGAAGUGCGAAAGUG-------GAAAGUAUGGGAAACAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUUAAAUGUUU .......(((((((((.((((.(...(.-------((((.....(....).)))--------).).).))))...)))))))))((((((((.....))))))))............... ( -29.70) >DroAna_CAF1 36309 113 + 1 UGCUUUAUGGUCAUGAAGUGCGAAAGUG-------GAAAAUAUGAAAAACAUUUUGUACCCUUGAAUGGCACAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUU .((....(((((((((.((((.((((.(-------(..((.(((.....))).))...)))))...).))))...)))))))))((((((((.....))))))))..))........... ( -32.40) >consensus UGUUUUAUGGUCAUGAAGUGCGAAAGUG_______GAAAGUAUGGGAAACAUUU________UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUU .((((((.((((((((.((((.(............(((.((.......)).)))............).))))...))))))))(((((((((.....)))))))))....)))))).... (-26.22 = -26.05 + -0.17)


| Location | 19,272,745 – 19,272,850 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19272745 105 - 22407834 AAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGUUGCCCAUACUUUC-------CACUUUCACACUUCAUGACCAUAAAACA .((((((((.....(((((((((.....)))))))))(((.(((...))))))....)--------)))))))............-------............................ ( -21.00) >DroSec_CAF1 13978 112 - 1 AAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGCUUCCCAUACUUUCCACUUUCCACUUUCGCACUUCAUGACCAUAAAACA .........((((((((((((((.....)))))))))(((.(((...)))))).....--------...))))).............................................. ( -19.90) >DroSim_CAF1 20858 105 - 1 AAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGCUUCCCAUACUUUC-------CACUUUCGCACUUCAUGACCAUAAAACA .........((((((((((((((.....)))))))))(((.(((...)))))).....--------...)))))...........-------............................ ( -19.90) >DroEre_CAF1 20730 105 - 1 AAGCAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUCGGCCAUAACUUAUGCCAUUCA--------AAAUGCUGCCCAUACUUUC-------CACUUUCGCACUUCAUGACCAUAAAACA .((((((((.....((((((.((((...((((((...))))))...)))))))))).)--------)))))))............-------............................ ( -23.00) >DroYak_CAF1 20021 105 - 1 AAACAUUUAAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGUUUCCCAUACUUUC-------CACUUUCGCACUUCAUGACCAUAAAACA ((((((((......(((((((((.....)))))))))(((.(((...)))))).....--------))))))))...........-------............................ ( -20.10) >DroAna_CAF1 36309 113 - 1 AAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUGUGCCAUUCAAGGGUACAAAAUGUUUUUCAUAUUUUC-------CACUUUCGCACUUCAUGACCAUAAAGCA (((((((((..((.(((((((((.....))))))))).))........(((((.......))))))))))))))...........-------............................ ( -27.10) >consensus AAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA________AAAUGCUUCCCAUACUUUC_______CACUUUCGCACUUCAUGACCAUAAAACA .(((((((......((((((.((((...((((((...))))))...))))))))))..........)))))))............................................... (-18.21 = -18.24 + 0.03)

| Location | 19,272,778 – 19,272,890 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -23.77 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
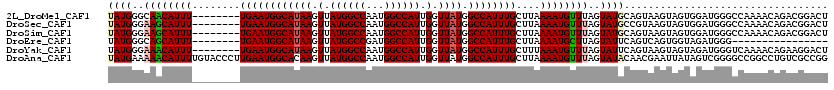
>2L_DroMel_CAF1 19272778 112 + 22407834 UAUGGGCAACAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCAGUAAGUAGUGGAUGGGCCAAAACAGACGGACU ....(((..(((((--------.(((((((((((.(.((((((...)))))).).)))).)))))))(((((...(((.......))).)))))...))))).))).............. ( -32.00) >DroSec_CAF1 14018 112 + 1 UAUGGGAAGCAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCCGUAAGUAGUGGAUGGGCCAAAACAGACGGACU ((((..((((((((--------((((((((....))))).((.(((((((((.....))))))))).)))))))))))))..))))((((..((..(((.....)))..))..))))... ( -37.60) >DroSim_CAF1 20891 112 + 1 UAUGGGAAGCAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCAGUAAGUAGUGGAUGGGCCAAAACAGACGGACU ........((((.(--------((((((...(((((.(((((((.(((((...))))))))))))..)))))...))))))).)))).((..((..(((.....)))..))..))..... ( -32.60) >DroEre_CAF1 20763 96 + 1 UAUGGGCAGCAUUU--------UGAAUGGCAUAAGUUAUGGCCGAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGCUUAGUAUUCAGUCAGUGGUAGAUGGG---------------- (((..((.((...(--------((((((.(.(((((...(((.(((((((((.....))))))))).)))......)))))))))))))...)).))..)))..---------------- ( -28.50) >DroYak_CAF1 20054 112 + 1 UAUGGGAAACAUUU--------UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUUAAAUGUUUAGUAUUCAGUAAGUAGUAGAUGGGUCAAAACAGAAGGACU ....(....).(((--------((..((((.....(((.(((.(((((((((.....))))))))).))).)))..(((((.((((.....)))).)))))..))))...)))))..... ( -28.40) >DroAna_CAF1 36342 120 + 1 UAUGAAAAACAUUUUGUACCCUUGAAUGGCACAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUACAACGAAUUAUAGUCGGGGCCGGCCUGUCGCCGG ((((..((((((((((.........(((((....)))))(((.(((((((((.....))))))))).)))))))))))))..))))(..(((.......)))..)(((((.....))))) ( -36.30) >consensus UAUGGGAAACAUUU________UGAAUGGCAUAAGUUAUGGCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCAGUAAGUAGUAGAUGGGCCAAAACAGACGGACU ((((..((((((((........((((((((((((.(.((((((...)))))).).)))).))))))))....))))))))..)))).................................. (-23.77 = -24.13 + 0.36)
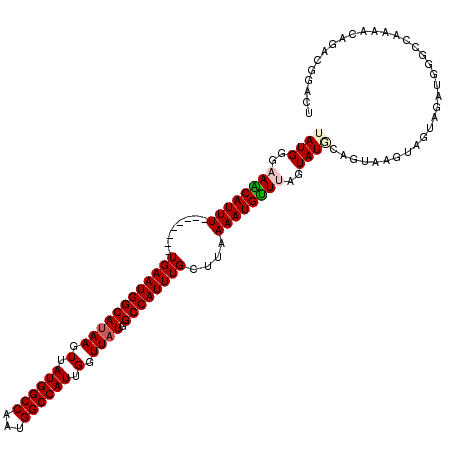
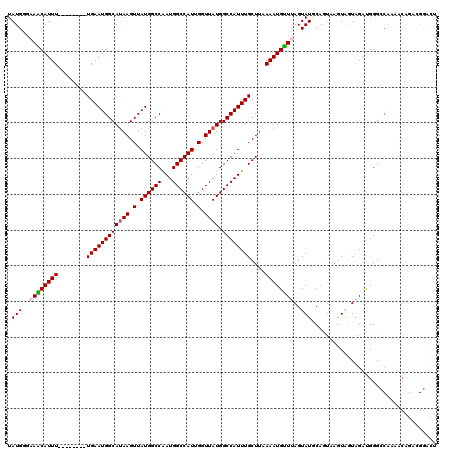
| Location | 19,272,778 – 19,272,890 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19272778 112 - 22407834 AGUCCGUCUGUUUUGGCCCAUCCACUACUUACUGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGUUGCCCAUA ....(((..(((.(((((..............(((................)))(((((((((.....)))))))))))))).)))..))).......--------.............. ( -25.59) >DroSec_CAF1 14018 112 - 1 AGUCCGUCUGUUUUGGCCCAUCCACUACUUACGGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGCUUCCCAUA .(.((((..((..(((.....)))..))..)))))..............((((((((((((((.....)))))))))(((.(((...)))))).....--------...)))))...... ( -29.30) >DroSim_CAF1 20891 112 - 1 AGUCCGUCUGUUUUGGCCCAUCCACUACUUACUGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGCUUCCCAUA ....(((..(((.(((((..............(((................)))(((((((((.....)))))))))))))).)))..))).......--------.............. ( -25.59) >DroEre_CAF1 20763 96 - 1 ----------------CCCAUCUACCACUGACUGAAUACUAAGCAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUCGGCCAUAACUUAUGCCAUUCA--------AAAUGCUGCCCAUA ----------------.........................((((((((.....((((((.((((...((((((...))))))...)))))))))).)--------)))))))....... ( -23.00) >DroYak_CAF1 20054 112 - 1 AGUCCUUCUGUUUUGACCCAUCUACUACUUACUGAAUACUAAACAUUUAAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA--------AAAUGUUUCCCAUA ........................................((((((((......(((((((((.....)))))))))(((.(((...)))))).....--------))))))))...... ( -20.30) >DroAna_CAF1 36342 120 - 1 CCGGCGACAGGCCGGCCCCGACUAUAAUUCGUUGUAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUGUGCCAUUCAAGGGUACAAAAUGUUUUUCAUA (((((.....)))))...((((........))))......(((((((((..((.(((((((((.....))))))))).))........(((((.......))))))))))))))...... ( -37.70) >consensus AGUCCGUCUGUUUUGGCCCAUCCACUACUUACUGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGCCAUAACUUAUGCCAUUCA________AAAUGCUUCCCAUA .........................................(((((((......((((((.((((...((((((...))))))...))))))))))..........)))))))....... (-18.21 = -18.24 + 0.03)
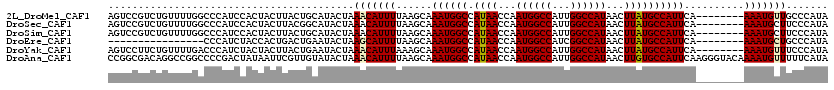


| Location | 19,272,810 – 19,272,915 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -18.53 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19272810 105 + 22407834 GCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCAGUAAGUAGUGGAUGGGCCAAAACAGACGGACUGAGUCCAAGGAAUAACUUAUU---------------UGCA ((.(((((((((.....))))))))).))...............((((((((((.((...(((((((...(....)...)).)))))....)).))))).)---------------)))) ( -30.10) >DroSec_CAF1 14050 105 + 1 GCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCCGUAAGUAGUGGAUGGGCCAAAACAGACGGACUGAGUCCAAGGAAUAACUUAUU---------------UGCA ((.(((((((((.....))))))))).))........((((((...((((..((..(((.....)))..))..))))))))))...(((......)))...---------------.... ( -29.00) >DroSim_CAF1 20923 105 + 1 GCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCAGUAAGUAGUGGAUGGGCCAAAACAGACGGACUGAGUCCAAGGAAUAACUUAUU---------------UGCA ((.(((((((((.....))))))))).))...............((((((((((.((...(((((((...(....)...)).)))))....)).))))).)---------------)))) ( -30.10) >DroEre_CAF1 20795 85 + 1 GCCGAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGCUUAGUAUUCAGUCAGUGGUAGAUGGG--------------------CCAAGGAAUAACUUGUU---------------UGCA ((.(((((((((.....))))))))).))............(((..(((((..((((......)--------------------)))..)).....)))..---------------))). ( -25.20) >DroYak_CAF1 20086 105 + 1 GCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUUAAAUGUUUAGUAUUCAGUAAGUAGUAGAUGGGUCAAAACAGAAGGACUCAGUCCAAGGAAUAACUUAUU---------------UGCA ((.(((((((((.....))))))))).))...................(((((((((...((((((..........)))))).((....))...)).))))---------------))). ( -28.00) >DroAna_CAF1 36382 120 + 1 GCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUACAACGAAUUAUAGUCGGGGCCGGCCUGUCGCCGGGAGGCGGAGGAAUAACUUAUCGCAGUAGAGCUUUUCGGGG ((.(((((((((.....))))))))).))......((...(((.(((..(((......((((....))))((..((((....))))..)).........)))..)))..)))...))... ( -37.30) >consensus GCCAAUGGCCAUUGGUUAUGGCCAUUUGCUUAAAAUGUUUAGUAUGCAGUAAGUAGUAGAUGGGCCAAAACAGACGGACUGAGUCCAAGGAAUAACUUAUU_______________UGCA ((.(((((((((.....))))))))).)).......(((((.(((.......))).)))))..............(((.....)))(((......)))...................... (-18.53 = -18.60 + 0.07)
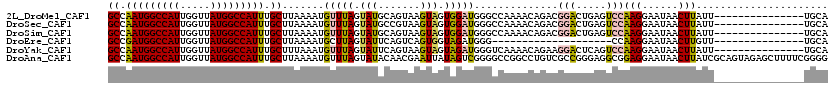

| Location | 19,272,810 – 19,272,915 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -14.75 |
| Energy contribution | -15.34 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19272810 105 - 22407834 UGCA---------------AAUAAGUUAUUCCUUGGACUCAGUCCGUCUGUUUUGGCCCAUCCACUACUUACUGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGC ((((---------------..(((((........((((...))))(((......))).........))))).))))...............((.(((((((((.....))))))))).)) ( -25.90) >DroSec_CAF1 14050 105 - 1 UGCA---------------AAUAAGUUAUUCCUUGGACUCAGUCCGUCUGUUUUGGCCCAUCCACUACUUACGGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGC (((.---------------....((((........))))..(.((((..((..(((.....)))..))..)))))................)))(((((((((.....)))))))))... ( -25.70) >DroSim_CAF1 20923 105 - 1 UGCA---------------AAUAAGUUAUUCCUUGGACUCAGUCCGUCUGUUUUGGCCCAUCCACUACUUACUGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGC ((((---------------..(((((........((((...))))(((......))).........))))).))))...............((.(((((((((.....))))))))).)) ( -25.90) >DroEre_CAF1 20795 85 - 1 UGCA---------------AACAAGUUAUUCCUUGG--------------------CCCAUCUACCACUGACUGAAUACUAAGCAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUCGGC (((.---------------......(((.((..(((--------------------........)))..)).))).......)))......((..((((((((.....))))))))..)) ( -19.54) >DroYak_CAF1 20086 105 - 1 UGCA---------------AAUAAGUUAUUCCUUGGACUGAGUCCUUCUGUUUUGACCCAUCUACUACUUACUGAAUACUAAACAUUUAAAGCAAAUGGCCAUAACCAAUGGCCAUUGGC ....---------------....(((.((((...((((...)))).((......)).................)))))))...........((.(((((((((.....))))))))).)) ( -20.20) >DroAna_CAF1 36382 120 - 1 CCCCGAAAAGCUCUACUGCGAUAAGUUAUUCCUCCGCCUCCCGGCGACAGGCCGGCCCCGACUAUAAUUCGUUGUAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGC ................(((((..((((((.....((....(((((.....)))))...))...))))))..)))))...............((.(((((((((.....))))))))).)) ( -31.30) >consensus UGCA_______________AAUAAGUUAUUCCUUGGACUCAGUCCGUCUGUUUUGGCCCAUCCACUACUUACUGCAUACUAAACAUUUUAAGCAAAUGGCCAUAACCAAUGGCCAUUGGC .....................(((((.......(((...(((..(....)..)))..)))......)))))....................((.(((((((((.....))))))))).)) (-14.75 = -15.34 + 0.59)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:41 2006