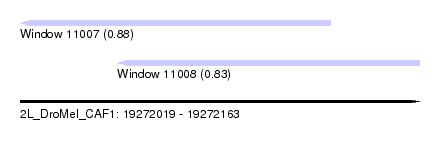
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,272,019 – 19,272,163 |
| Length | 144 |
| Max. P | 0.880735 |

| Location | 19,272,019 – 19,272,131 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -22.13 |
| Energy contribution | -23.72 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
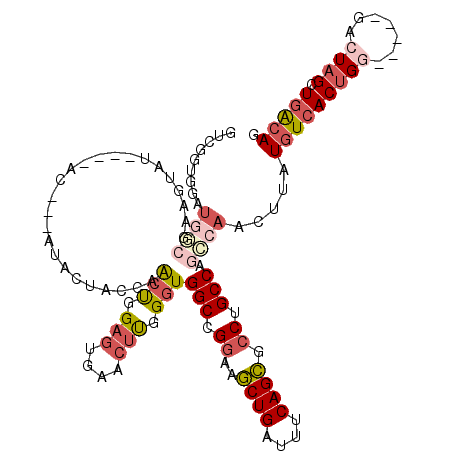
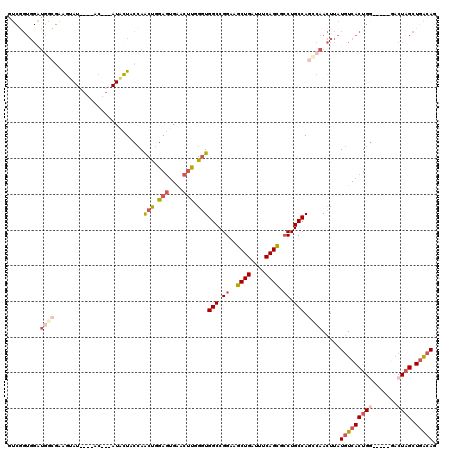
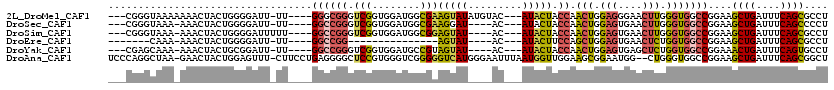
>2L_DroMel_CAF1 19272019 112 - 22407834 GUCGGUGGAUGGCGAAGUAUAUGUAC---AUACUACCAACUGGAGGGAACUUGGGUGGCCGGAAGCUGAUUUCAGCGCCUGCCAGCCAACUUAUGUCACUGG-----GACUAGCUGACAG ((((((((.((((..(((((......---)))))....(((.(((....))).)))(((.((..((((....)))).)).))).)))).))...(((.....-----)))..)))))).. ( -39.00) >DroSec_CAF1 13258 108 - 1 GUCGGUGGAUGGCGAAGGAU----AC---AUACUACCAACUGGAGUGAACUUGGGUGGCCGGAAGCUGAUUUCAGCCCCUGCCAGCCAACUUAUGUCACUGG-----GUCUAGCUGACAG (((((((((((.(....)..----.)---)).)))))..(((((((((..((((.((((.((..((((....)))).)).)))).))))......))))...-----.)))))..))).. ( -41.10) >DroSim_CAF1 20140 108 - 1 GUCGGUGGAUGGCGGAGUAU----AC---AUACUACCAACUGGAGUGAACUUGGGUGGCCGGAAGCUGAUUUCAGCGCCUGCCAGCCAACUUAUGUCACUGG-----GACUAGCUGACAG ((((((((.((((((((((.----..---.)))).)).(((.(((....))).)))(((.((..((((....)))).)).))).)))).))...(((.....-----)))..)))))).. ( -40.60) >DroEre_CAF1 20007 93 - 1 ---------------AGUAU----AC---AUACUUCCAGCUGGAGUGAACUCUGGUGGCCGGAAGCUGAUUUCAGCGCCUGCCAGGCAAUUUAUGUCACUGG-----GAGUAGCUGACAG ---------------.((..----..---.((((((((((..(((....)))..)((((.((..((((....)))).)).))))((((.....)))).))))-----)))))....)).. ( -38.40) >DroYak_CAF1 19279 108 - 1 GUCGGUGGAUGCCGUAGUAU----AC---AUACUACCAACUGGAGUGAGCUCUGGUGGCCGGAAACUGAUUUCAGUGCCUGCCAGGCAACUUAUGUCACUGG-----CACUAGCUGACAG ((((((...((((((((((.----..---.))))))..((..(((....)))..))(((.((..((((....)))).)).))).((((.....))))...))-----))...)))))).. ( -42.40) >DroAna_CAF1 35474 118 - 1 CUCCGUGGGUCGGGGGUCAUGGGAAUUUAAUGGUUGGAAGCGGAAUGG--CUGGGUGGCCGGAAGCUGAUUUCAGCGGCUGCCAAGCAACUUAUGUCACUGACUUGUAACUUGCUUGAAA .(((((((.(....).)))))))..(((((.(((((.((((((....(--((.((..(((....((((....)))))))..)).))).((....))..))).))).)))))...))))). ( -38.60) >consensus GUCGGUGGAUGGCGAAGUAU____AC___AUACUACCAACUGGAGUGAACUUGGGUGGCCGGAAGCUGAUUUCAGCGCCUGCCAGCCAACUUAUGUCACUGG_____GACUAGCUGACAG .........((((.........................(((.(((....))).)))(((.((..((((....)))).)).))).)))).....(((((((((.......)))).))))). (-22.13 = -23.72 + 1.59)

| Location | 19,272,054 – 19,272,163 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Mean single sequence MFE | -38.11 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19272054 109 - 22407834 ---CGGGUAAAAAAACUACUGGGGAUU-UU----GGGCGGGUCGGUGGAUGGCGAAGUAUAUGUAC---AUACUACCAACUGGAGGGAACUUGGGUGGCCGGAAGCUGAUUUCAGCGCCU ---(.((((.......)))).).....-..----((((.((((((((((((..............)---)).))))).(((.(((....))).)))))))....((((....)))))))) ( -35.34) >DroSec_CAF1 13293 104 - 1 ---CGGGUAAA-AAACUACUGGGGAUU-UU----GGCCGGGUCGGUGGAUGGCGAAGGAU----AC---AUACUACCAACUGGAGUGAACUUGGGUGGCCGGAAGCUGAUUUCAGCCCCU ---..((((..-....))))((((.((-((----((((.....((((((((.(....)..----.)---)).))))).(((.(((....))).)))))))))))((((....)))))))) ( -40.20) >DroSim_CAF1 20175 105 - 1 ---CGGGUAAA-AAACUACUGGGGAUUUUU----GGCCGGGUCGGUGGAUGGCGGAGUAU----AC---AUACUACCAACUGGAGUGAACUUGGGUGGCCGGAAGCUGAUUUCAGCGCCU ---(((.((((-((.((.....)).)))))----).)))((((((((((((.........----.)---)).))))).(((.(((....))).)))))))((..((((....)))).)). ( -36.60) >DroEre_CAF1 20042 85 - 1 -------CAAA-AAACUACUGGGGAUU-UU----GGCCGG---------------AGUAU----AC---AUACUUCCAGCUGGAGUGAACUCUGGUGGCCGGAAGCUGAUUUCAGCGCCU -------....-.........(((.((-((----((((((---------------((((.----..---.))))))..((..(((....)))..))))))))))((((....)))).))) ( -33.50) >DroYak_CAF1 19314 104 - 1 ---CGAGCAAA-AAACUACUGCGGAUU-UU----GGCCGGGUCGGUGGAUGCCGUAGUAU----AC---AUACUACCAACUGGAGUGAGCUCUGGUGGCCGGAAACUGAUUUCAGUGCCU ---.((((...-....((((((((...-((----.((((...)))).))..)))))))).----..---.((((.((....)))))).))))....(((.....((((....))))))). ( -35.00) >DroAna_CAF1 35514 116 - 1 UCCCAGGCUAA-GAACUACUGGAGUUU-CUUCCUGAGGGGCUCCGUGGGUCGGGGGUCAUGGGAAUUUAAUGGUUGGAAGCGGAAUGG--CUGGGUGGCCGGAAGCUGAUUUCAGCGGCU (((((((((..-((.((((.(((((..-(((...)))..))))))))).))...)))).)))))........(((((((.(((..(((--((....)))))....))).))))))).... ( -48.00) >consensus ___CGGGCAAA_AAACUACUGGGGAUU_UU____GGCCGGGUCGGUGGAUGGCGAAGUAU____AC___AUACUACCAACUGGAGUGAACUUGGGUGGCCGGAAGCUGAUUUCAGCGCCU ..................................((((((.(((........)))(((((.........))))).)).(((.(((....))).)))))))....((((....)))).... (-15.28 = -15.50 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:34 2006