| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,269,088 – 19,269,189 |
| Length | 101 |
| Max. P | 0.725183 |

| Location | 19,269,088 – 19,269,189 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.06 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.80 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
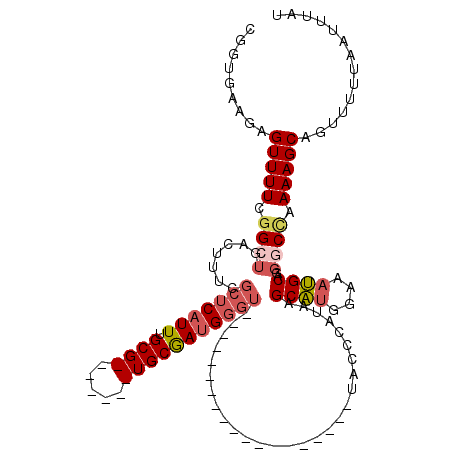
>2L_DroMel_CAF1 19269088 101 - 22407834 CGGUAAAGAGUUUUCGGCUGACUCUCGCUCAUUUGCG-------UGCGAUGGGUAUACCC------------AUACCCAUAAGCAUGGAAAUGCAGGGCCAAAAGCAGUUUUUAAUUUAU ...(((((((((((.(((((.....).(((((((.((-------(((.((((((((....------------))))))))..))))).))))).)))))).)))))..))))))...... ( -32.10) >DroSec_CAF1 10383 113 - 1 CGGUGAAGAGUUUUCGGCUGACUUUCGCUCAUUUGCG-------UGCGAUGGGUAUACCCACACCCAUACCCAUACCCAUAAGCAUGGAAAUGCAGGGCUAAAAGCAGUUUUUAAUUUAU .......(((((...(((((.(((((.(((((((.((-------(((.((((((((................))))))))..))))).))))).)).)...)))))))))...))))).. ( -30.89) >DroSim_CAF1 17207 113 - 1 CGGUGAAGAGUUUUCGGCUGACUUUCGCUCAUUUGCG-------UGCGAUGGGUAUACCCACACCCAUACCCGUACCCAUAAGCAUGGAAAUGCAGGGCCAAAAGCAGUUUUUAAUUUAU .......(((((...(((((.((((.((((...((.(-------((((((((((........))))))...))))).))...((((....)))).))))..)))))))))...))))).. ( -32.80) >DroEre_CAF1 17241 87 - 1 UGGUGAAGAGUUUUCGGAUGACUUUCGCUCAUUUGCG-------UGCGAUGGGU--------------------------GAGCAUGGAAAUGCAGGGCCAAAAGCAGUUUUUAAUUUAU ...(((((((((((.((....((((((((((((.(..-------..))))))))--------------------------))((((....))))))).)).)))))..))))))...... ( -21.40) >DroYak_CAF1 16313 87 - 1 UGGUGCUGAGUUUUCCGAUGACUUUCGCUCAUUUGCG-------UGCGAUGGUU--------------------------AAGCACGGAAAUGCAGGGCCAAAAGCAGUUUUUCAUUUAU ((((.(((...((((((.(((((.((((.(......)-------.)))).))))--------------------------)....))))))..))).))))................... ( -27.60) >DroAna_CAF1 32665 98 - 1 --CUGGAGAGUUUUCGGCUGACUCCCGCUCAUUUGCGAAGCUUUUGCAAAGCGG------------------GCAAGGGCAAGCGUGUAAACGCA--GCCAAAAGCAGUUUUUAAUUUAU --.......(((((.((((..((((((((...((((((.....)))))))))))------------------)..)).....((((....)))))--))).))))).............. ( -30.80) >consensus CGGUGAAGAGUUUUCGGCUGACUUUCGCUCAUUUGCG_______UGCGAUGGGU___________________UACCCAUAAGCAUGGAAAUGCAGGGCCAAAAGCAGUUUUUAAUUUAU .........(((((.((((.......(((((((.((((.....)))))))))))............................((((....))))..)))).))))).............. (-16.62 = -17.80 + 1.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:30 2006