
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,266,778 – 19,266,915 |
| Length | 137 |
| Max. P | 0.694613 |

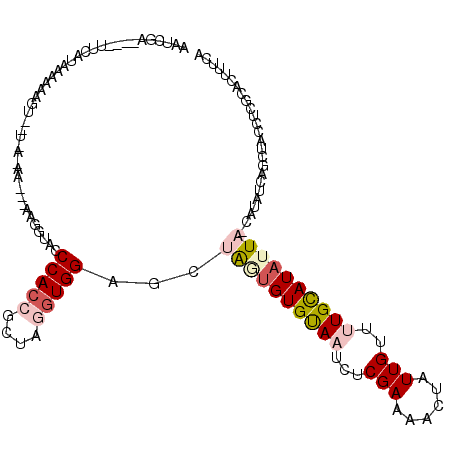
| Location | 19,266,778 – 19,266,884 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.64 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -8.70 |
| Energy contribution | -10.06 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19266778 106 + 22407834 AAUCGGAUUCUUCAUUCAUAAG--------------GUACCCCACCGCUAGGUGGAGCUGAUGUGUAAUCUCGAUAACUAUUGUUUUGUAUAUUAGCAUAUCAGCUACCUCGCACUUUCA ....((((.....))))...((--------------((...(((((....)))))((((((((((((((..(((.(((....))))))...))).))))))))))))))).......... ( -29.60) >DroSec_CAF1 8307 80 + 1 ----------------------------------------ACCACCGCUAGGUGGCGCUAGUGUGUAAUCUCGAAAACUAUUGUUUUGUAUAUUAUCAUAUCAGCUACCUCGCACAUUCA ----------------------------------------......((.(((((((....(((((((((....(((((....)))))....)))).)))))..))))))).))....... ( -21.00) >DroSim_CAF1 15046 118 + 1 AAUCGAAAUAUUCAUUAAAAAGUUGUAAAAAC--AAGUUACCCACAGCUAGGUGGCGCUAGUGUGUAAUCUCGAAAAUCAUUGUUUUGCAUUUUAGCAUAUCAGCUACCUCGCACUUUCA ....((((...........((.((((....))--)).)).......((.((((((((((((.((((((...(((......)))..)))))).)))))......))))))).))..)))). ( -27.20) >DroEre_CAF1 14984 105 + 1 AAUGAACAAGUUCAUAAAAGAGUUUUGACAAUUGAAUGCACCCACCGCUAGGUGGAGCUAUUGUGCAAUCUCGAAAACUGUUGCGCUGCAUAUUU---------------CGCAGUUUCA .(((((....)))))..((.((((((((.....((.((((((((((....))))).......))))).)))).)))))).))..(((((......---------------.))))).... ( -26.91) >DroYak_CAF1 14016 111 + 1 GAUGCA----UUAAUAAAAUUGUAAUGACAAACGAAUGUACCCAAAGCUAGGUGGGGCUACUGUGUAAUAUCGAACACUAUUGCUGUGCAUAUUU-----UCAGCUACCUAGCACUUUCA ..((((----((.......((((....))))...))))))......((((((((((((....((((........))))....))).((.......-----.)).)))))))))....... ( -23.70) >consensus AAUCGA____UUCAUAAAAAAGU__U_A_AA___AAGGUACCCACCGCUAGGUGGAGCUAGUGUGUAAUCUCGAAAACUAUUGUUUUGCAUAUUA_CAUAUCAGCUACCUCGCACUUUCA .........................................(((((....)))))...((((((((((...(((......)))..))))))))))......................... ( -8.70 = -10.06 + 1.36)

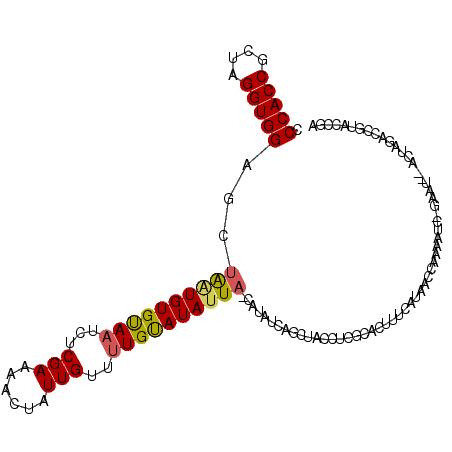
| Location | 19,266,804 – 19,266,915 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 68.84 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19266804 111 + 22407834 CCCACCGCUAGGUGGAGCUGAUGUGUAAUCUCGAUAACUAUUGUUUUGUAUAUUAGCAUAUCAGCUACCUCGCACUUUCAUAAUCAAAGUC-GAAU-AACUAGUCCGUACCGA .(((((....)))))((((((((((((((..(((.(((....))))))...))).)))))))))))...(((.(((((.......))))))-))..-................ ( -30.00) >DroSec_CAF1 8307 112 + 1 ACCACCGCUAGGUGGCGCUAGUGUGUAAUCUCGAAAACUAUUGUUUUGUAUAUUAUCAUAUCAGCUACCUCGCACAUUCAUAACUAUAAUAUUAAUU-UCUUGCCCGAACCGA ......((.(((((((....(((((((((....(((((....)))))....)))).)))))..))))))).))........................-............... ( -21.00) >DroEre_CAF1 15024 95 + 1 CCCACCGCUAGGUGGAGCUAUUGUGCAAUCUCGAAAACUGUUGCGCUGCAUAUUU---------------CGCAGUUUCACAAGCUG-AUC-GAAA-UACUGCACUAUACCGA .(((((....))))).......(((((((..........)))))))(((((((((---------------(((((((.....)))))-..)-))))-)).))))......... ( -28.50) >consensus CCCACCGCUAGGUGGAGCUAAUGUGUAAUCUCGAAAACUAUUGUUUUGUAUAUUA_CAUAUCAGCUACCUCGCACUUUCAUAACCAAAAUC_GAAU__ACUAGACCGUACCGA .(((((....)))))...((((((((((...(((......)))..)))))))))).......................................................... (-12.24 = -12.47 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:26 2006