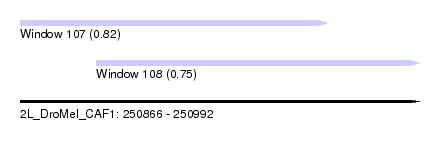
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 250,866 – 250,992 |
| Length | 126 |
| Max. P | 0.817212 |

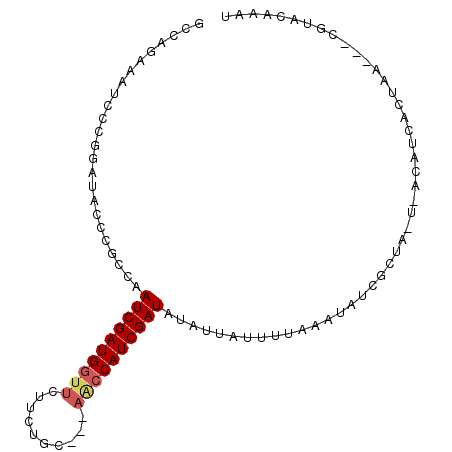
| Location | 250,866 – 250,963 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -10.02 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 250866 97 + 22407834 GCCAGAAGUCCCGGAUACCCGCCAAUCGAUGGUUCUUCUUUUUAUGCCAUCGAUAUAUUAUUUUAAAUAUCGCUACUGACAUCACUAA---UGUACAAAU ..(((.(((..(((....)))...(((((((((............))))))))).................))).)))((((.....)---)))...... ( -19.20) >DroSec_CAF1 42614 91 + 1 GCCAGAAAUCCGAGAUACCCGCCAAUCGAUGGUUCUUCUGC---AACCAUCGAUAUAUUAUUUUAAAUAUCUCUA---ACAUCAUUGA---CGUACAAAU ...........((((((.......(((((((((((....).---)))))))))).((......))..))))))..---..........---......... ( -17.50) >DroSim_CAF1 44258 91 + 1 GCCAGAAAUCCGGGAUACCCGCCAAUCGAUGGUUCUUCUGC---AACCAUCGAUAUAUUAUUUUAAAUAUCUCUA---ACAUCACUAA---CGUACAAAU ...(((....((((...))))...(((((((((((....).---)))))))))).................))).---.....((...---.))...... ( -18.70) >DroEre_CAF1 44561 84 + 1 GCCAGAAAUCCCGGAUACCU-CCAAUCGAUGGUUGCAGAAC---AACCAUCGAUAUUGUAUGUUCAGUAUCGCUAGUAA------------CGCACCAAU ((..........(((....)-))....((((((((.....)---)))))))(((((((......)))))))........------------.))...... ( -22.20) >DroYak_CAF1 46097 96 + 1 GCCAGAAAUCCCGGAUAACUGCCAAUCGAUGGAUGUU-CGC---AAGCAUCGAUAUUAUAUUUUCAGUAUCGCUACUAACAUCAGUAACGACGUACCAAU ....((((....((.......)).(((((((..((..-..)---)..))))))).......)))).((((((.((((......)))).)))..))).... ( -16.30) >consensus GCCAGAAAUCCCGGAUACCCGCCAAUCGAUGGUUCUUCUGC___AACCAUCGAUAUAUUAUUUUAAAUAUCGCUA_U_ACAUCACUAA___CGUACAAAU ........................((((((((((..........)))))))))).............................................. (-10.02 = -10.50 + 0.48)

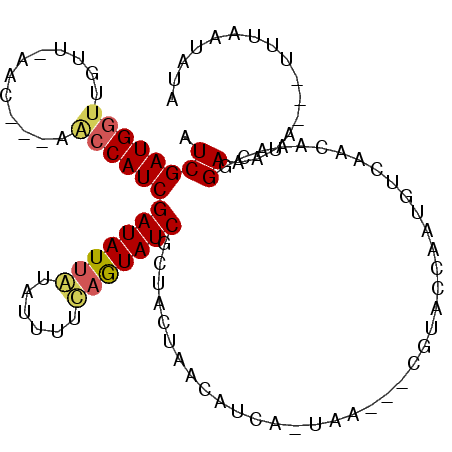
| Location | 250,890 – 250,992 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 68.30 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -8.92 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 250890 102 + 22407834 AUCGAUGGUUCUUCUUUUUAUGCCAUCGAUAUAUUAUUUUAAAUAUCGCUACUGACAUCACUAA---UGUACAAAUGACAACAAACGGAAAUAUUAUUUAACAAA (((((((((............)))))))))........(((((((......(((((((.....)---))).....((....))..)))......))))))).... ( -14.80) >DroEre_CAF1 44584 87 + 1 AUCGAUGGUUGCAGAAC---AACCAUCGAUAUUGUAUGUUCAGUAUCGCUAGUAA------------CGCACCAAUGUUAACAAAAGGAAAUA---UUUAAUAUA (((((((((((.....)---))))))))))....((((((.(((((..((..(((------------((......)))))......))..)))---)).)))))) ( -21.50) >DroYak_CAF1 46121 98 + 1 AUCGAUGGAUGUU-CGC---AAGCAUCGAUAUUAUAUUUUCAGUAUCGCUACUAACAUCAGUAACGACGUACCAAUGUCAACAAACGGAAAUC---UUAAAUAUA .(((((((((((.-...---..)))))((((((........))))))........))))......(((((....)))))........))....---......... ( -17.70) >consensus AUCGAUGGUUGUU_AAC___AACCAUCGAUAUUAUAUUUUCAGUAUCGCUACUAACAUCA_UAA___CGUACCAAUGUCAACAAACGGAAAUA___UUUAAUAUA .((((((((............))))))(((((((......)))))))........................................))................ ( -8.92 = -9.70 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:59 2006