
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
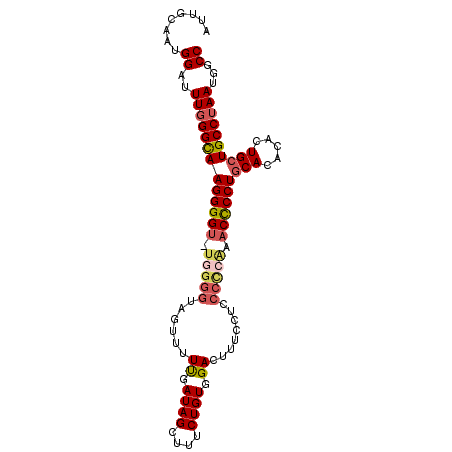
| Location | 19,254,858 – 19,254,955 |
| Length | 97 |
| Max. P | 0.691776 |

| Location | 19,254,858 – 19,254,955 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -23.18 |
| Energy contribution | -24.49 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
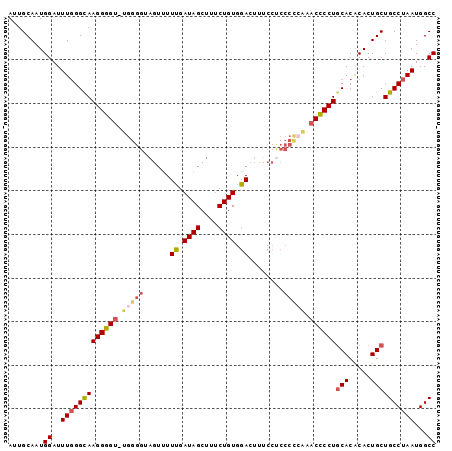
>2L_DroMel_CAF1 19254858 97 + 22407834 AUUGCAAUGGAUUUGGGCAAGGGGU-UGGGGUAGUAUUUGAUAGCUUUCUGUGGACUUUCCUCCCCCAAACCCCUUCACGCACUGCUGCCUAAUGGCC ........((..(((((((((((((-(((((.((...((.((((....)))).)).....)).)))).))))))))...((...)).))))))...)) ( -31.30) >DroSim_CAF1 2029 97 + 1 AUUGCAAUGGAUUUCGGCAAGGGGU-UGGGGUAGUUUUUGAUAGCUUGCUGUGGACUUUCCUCCCCCAAACCCCUGCACACACUGCUGCCUAAUGGCC ...(((.((.......(((.(((((-(((((.((...((.((((....)))).)).....)).)))).)))))))))...)).))).(((....))). ( -33.70) >DroEre_CAF1 3013 83 + 1 AUUGCAAUGGAUUUGGGUAAGGGGUUUGGGGUAGUUUUUGAUAGCUUUCUGUGGA---------------CUCCUGCACACACUGCUGCCUAAUGGCC ........((..(((((((((((((..(.((.((((......))))..)).)..)---------------)))))(((.....))))))))))...)) ( -24.40) >DroYak_CAF1 2065 97 + 1 AUUGCAAUGGAUUUGGGUAAGGGGU-UGGGGAAGUUUUCGAUAGCUUUCUGUGGACUUCCCCCCCUGGGACUCCUGCACACACUGCUGCCUAAUGGCC ...(((.((.....((((..((((.-.(((((((((..(((......)).)..)))))))))))))...)).))......)).))).(((....))). ( -34.50) >consensus AUUGCAAUGGAUUUGGGCAAGGGGU_UGGGGUAGUUUUUGAUAGCUUUCUGUGGACUUUCCUCCCCCAAACCCCUGCACACACUGCUGCCUAAUGGCC ........((..(((((((((((((.(((((......((.((((....)))).))........))))).))))))(((.....))))))))))...)) (-23.18 = -24.49 + 1.31)

| Location | 19,254,858 – 19,254,955 |
|---|---|
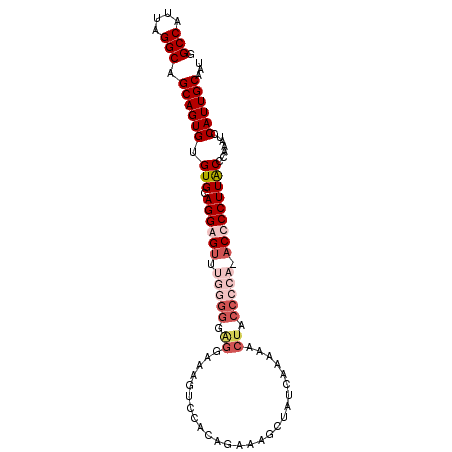
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -17.90 |
| Energy contribution | -20.77 |
| Covariance contribution | 2.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
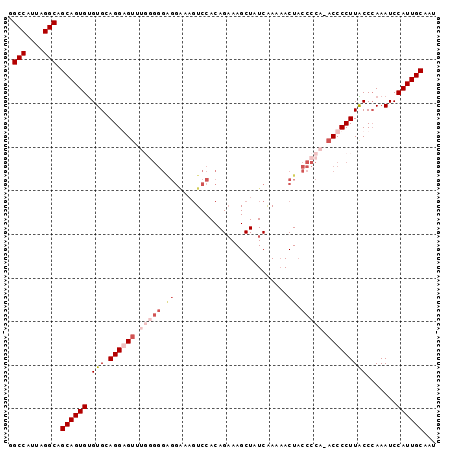
>2L_DroMel_CAF1 19254858 97 - 22407834 GGCCAUUAGGCAGCAGUGCGUGAAGGGGUUUGGGGGAGGAAAGUCCACAGAAAGCUAUCAAAUACUACCCCA-ACCCCUUGCCCAAAUCCAUUGCAAU .(((....))).((((((..((((((((((.((((..((.....))...((......))........)))))-)))))))...))....))))))... ( -33.90) >DroSim_CAF1 2029 97 - 1 GGCCAUUAGGCAGCAGUGUGUGCAGGGGUUUGGGGGAGGAAAGUCCACAGCAAGCUAUCAAAAACUACCCCA-ACCCCUUGCCGAAAUCCAUUGCAAU .(((....))).((((((((.(((((((.((((((..((.....))..((....))...........)))))-).))))))))).....))))))... ( -37.10) >DroEre_CAF1 3013 83 - 1 GGCCAUUAGGCAGCAGUGUGUGCAGGAG---------------UCCACAGAAAGCUAUCAAAAACUACCCCAAACCCCUUACCCAAAUCCAUUGCAAU .(((....))).((((((((((......---------------..))))((......))..............................))))))... ( -13.30) >DroYak_CAF1 2065 97 - 1 GGCCAUUAGGCAGCAGUGUGUGCAGGAGUCCCAGGGGGGGAAGUCCACAGAAAGCUAUCGAAAACUUCCCCA-ACCCCUUACCCAAAUCCAUUGCAAU .(((....))).((((((..(...((.((...((((((((((((.....((......))....)))))))..-.))))).))))..)..))))))... ( -33.10) >consensus GGCCAUUAGGCAGCAGUGUGUGCAGGAGUUUGGGGGAGGAAAGUCCACAGAAAGCUAUCAAAAACUACCCCA_ACCCCUUACCCAAAUCCAUUGCAAU .(((....))).((((((.(((.((((((.(((((.((..........................)).))))).))))))))).......))))))... (-17.90 = -20.77 + 2.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:12 2006