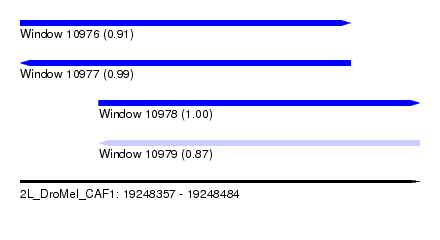
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,248,357 – 19,248,484 |
| Length | 127 |
| Max. P | 0.998989 |

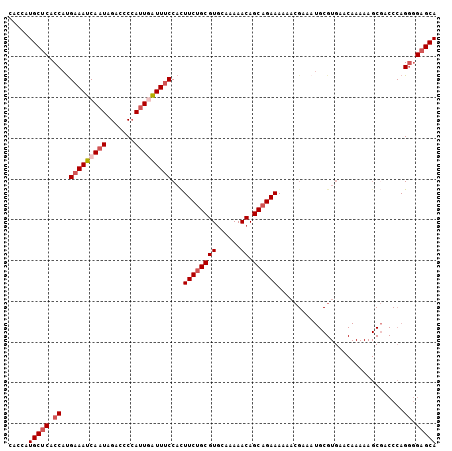
| Location | 19,248,357 – 19,248,462 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -22.10 |
| Energy contribution | -23.18 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910603 |
| Prediction | RNA |
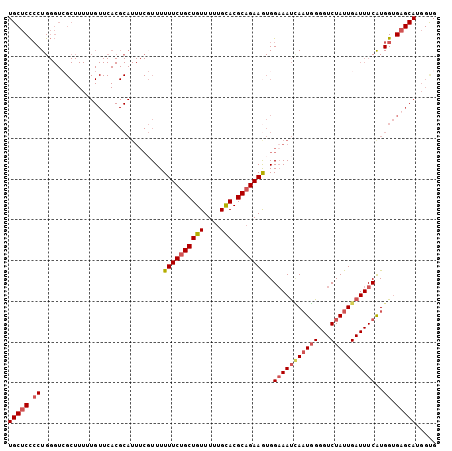
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19248357 105 + 22407834 UGCUCCCCUGGGUCGCUUUUUGUUCACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCAGAAGUGGAAAUUAAUGGGGUCUAUUGAUUUCAUGGUGAGCACGGUG .((.((....))..))....(((((((.(((.......((((((((((....))).)))))))..(((((((((((...)))))))))))))))))))))..... ( -32.20) >DroSec_CAF1 68264 105 + 1 UGCUCCCCUGGGUCGCUUUUUGUUCACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCUGAAGUGGAAAUCAAUGGGGUCUAUUGAUUUCGUGGUGAGCAUGGUG ....((....)).((((...(((((((.(((((((........(((((....))).)))))))))(((((((((((...)))))))))))...))))))).)))) ( -31.20) >DroSim_CAF1 69604 105 + 1 UGCUCCCCUGGGUCGCUUUUUGUUCACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCAGAAGUGGAAAUCAAUGGGGUCUAUUGAUUUCAUGGUGAGCAUGGUG ....((....)).((((...(((((((.(((.......((((((((((....))).)))))))..(((((((((((...))))))))))))))))))))).)))) ( -35.40) >DroEre_CAF1 67466 80 + 1 UGCUCUGCUG-------------------------UUUUUUCUGCUGUUUUUGCACGCAGAAAUGGAAACCAAUGGGGUCUAUUGAUUUCAUGGCGAGCAUUGUG (((((.((((-------------------------(((((((((((((....))).))))))...)))))..(((..(((....)))..)))))))))))..... ( -28.30) >DroYak_CAF1 69693 104 + 1 UGCUCCCCUGGGUAGCUUUUUGUUCACGCAUUUC-UUUUUUCUGCUGUUUUUGUACGCAGAAAUGGCAAUGACUCGGGUCUAUUGAUUUCAUGGUGUGCAUGGCG (((((....)))))(((...(((.(((.(((...-.......(((((((((((....)))))))))))..(((....)))..........)))))).))).))). ( -26.60) >consensus UGCUCCCCUGGGUCGCUUUUUGUUCACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCAGAAGUGGAAAUCAAUGGGGUCUAUUGAUUUCAUGGUGAGCAUGGUG (((((.((..............................((((((((((....))).)))))))..(((((((((((...)))))))))))..)).)))))..... (-22.10 = -23.18 + 1.08)

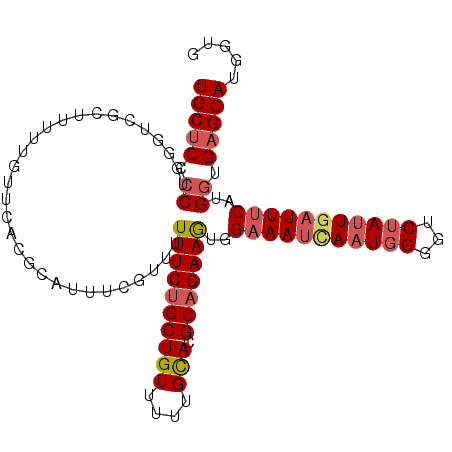
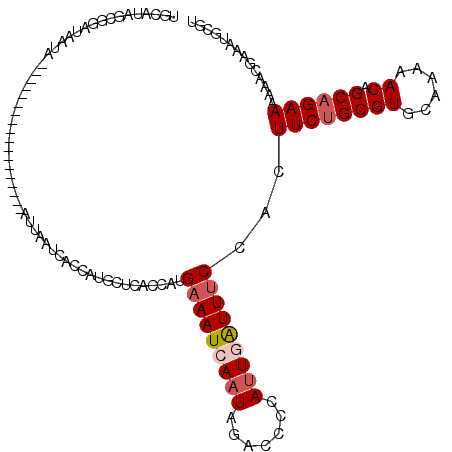
| Location | 19,248,357 – 19,248,462 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -18.30 |
| Energy contribution | -19.54 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19248357 105 - 22407834 CACCGUGCUCACCAUGAAAUCAAUAGACCCCAUUAAUUUCCACUUCUGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGUGAACAAAAAGCGACCCAGGGGAGCA .....(((((....((....))......(((....(((((...((((((((......)).)))))).....)))))((...........))......)))))))) ( -21.70) >DroSec_CAF1 68264 105 - 1 CACCAUGCUCACCACGAAAUCAAUAGACCCCAUUGAUUUCCACUUCAGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGUGAACAAAAAGCGACCCAGGGGAGCA (((((((((......(((((((((.......)))))))))......))))))........(((............))))))........((..((....)).)). ( -22.30) >DroSim_CAF1 69604 105 - 1 CACCAUGCUCACCAUGAAAUCAAUAGACCCCAUUGAUUUCCACUUCUGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGUGAACAAAAAGCGACCCAGGGGAGCA .....((.((((((((((((((((.......)))))))))...((((((((......)).))))))........))).)))).))....((..((....)).)). ( -26.70) >DroEre_CAF1 67466 80 - 1 CACAAUGCUCGCCAUGAAAUCAAUAGACCCCAUUGGUUUCCAUUUCUGCGUGCAAAAACAGCAGAAAAAA-------------------------CAGCAGAGCA .....(((((((...(((((((((.......)))))))))..(((((((((......)).)))))))...-------------------------..)).))))) ( -23.80) >DroYak_CAF1 69693 104 - 1 CGCCAUGCACACCAUGAAAUCAAUAGACCCGAGUCAUUGCCAUUUCUGCGUACAAAAACAGCAGAAAAAA-GAAAUGCGUGAACAAAAAGCUACCCAGGGGAGCA ...((((((...........((((.(((....)))))))...(((((((((......)).)))))))...-....))))))........(((.((....))))). ( -25.30) >consensus CACCAUGCUCACCAUGAAAUCAAUAGACCCCAUUGAUUUCCACUUCUGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGUGAACAAAAAGCGACCCAGGGGAGCA .....(((((.((..(((((((((.......)))))))))...((((((((......)).))))))...............................)).))))) (-18.30 = -19.54 + 1.24)

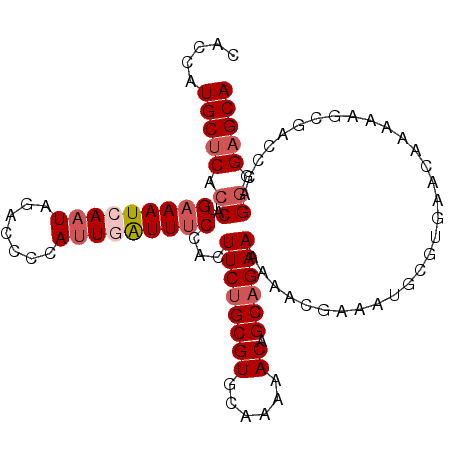
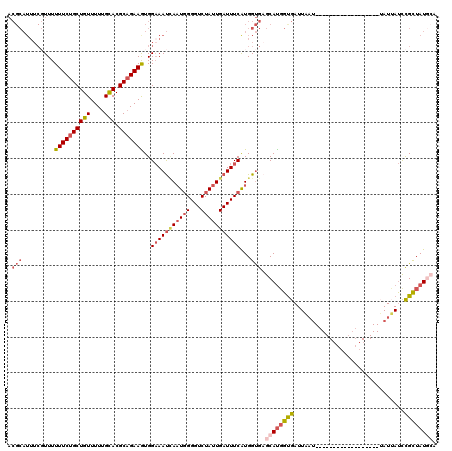
| Location | 19,248,382 – 19,248,484 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -18.32 |
| Energy contribution | -20.32 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19248382 102 + 22407834 ACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCAGAAGUGGAAAUUAAUGGGGUCUAUUGAUUUCAUGGUGAGCACGGUGAUUAAU------------------UAUUAUCCGUUAUGCA ..((((..((....(((((((((....))).))))))(((...((((.((..(((....)))..))))))...)))((((((....------------------.))))))))..)))). ( -29.30) >DroSec_CAF1 68289 102 + 1 ACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCUGAAGUGGAAAUCAAUGGGGUCUAUUGAUUUCGUGGUGAGCAUGGUGAUUAAU------------------UAUUAUUCGCUAUGCA .(((.........((((.(((((....))).)).))))..(((((((((((...))))))))))))))....(((((((((.((..------------------...)).))))))))). ( -32.00) >DroSim_CAF1 69629 102 + 1 ACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCAGAAGUGGAAAUCAAUGGGGUCUAUUGAUUUCAUGGUGAGCAUGGUGAUUAAU------------------UAUUAUUCGCUAUGCA .(((.........((((((((((....))).)))))))..(((((((((((...)))))))))))...))).(((((((((.((..------------------...)).))))))))). ( -36.10) >DroEre_CAF1 67476 106 + 1 ----------UUUUUUCUGCUGUUUUUGCACGCAGAAAUGGAAACCAAUGGGGUCUAUUGAUUUCAUGGCGAGCAUUGUGAUUAAUACAUUUCAC----ACAUUAAUAAUCUGCUAUAAU ----------...((((((((((....))).))))))).(((..(((.((..(((....)))..))))).......(((((..........))))----).........)))........ ( -24.10) >DroYak_CAF1 69718 119 + 1 ACGCAUUUC-UUUUUUCUGCUGUUUUUGUACGCAGAAAUGGCAAUGACUCGGGUCUAUUGAUUUCAUGGUGUGCAUGGCGAUUUUCACAUUACAUAUGUACAUUAAUAACCCGCUAUUAU ..((.....-.......(((((((((((....))))))))))).......((((.(((((((..((((.((((.(((..........)))))))))))...)))))))))))))...... ( -29.80) >consensus ACGCAUUUCGUUUUUUCUGCUGUUUUUGCACGCAGAAGUGGAAAUCAAUGGGGUCUAUUGAUUUCAUGGUGAGCAUGGUGAUUAAU__________________UAUUAUCCGCUAUGCA .(((.........((((((((((....))).)))))))..(((((((((((...)))))))))))...))).((((((((...............................)))))))). (-18.32 = -20.32 + 2.00)

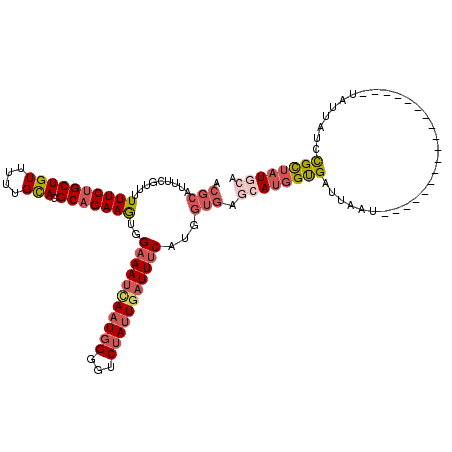
| Location | 19,248,382 – 19,248,484 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -10.42 |
| Energy contribution | -11.26 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19248382 102 - 22407834 UGCAUAACGGAUAAUA------------------AUUAAUCACCGUGCUCACCAUGAAAUCAAUAGACCCCAUUAAUUUCCACUUCUGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGU .((((...(((....(------------------((((((...((((.....))))...((....))....))))))))))..((((((((......)).))))))........)))).. ( -16.90) >DroSec_CAF1 68289 102 - 1 UGCAUAGCGAAUAAUA------------------AUUAAUCACCAUGCUCACCACGAAAUCAAUAGACCCCAUUGAUUUCCACUUCAGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGU .((((..((.......------------------..........(((((......(((((((((.......)))))))))......)))))((.......))........))..)))).. ( -19.30) >DroSim_CAF1 69629 102 - 1 UGCAUAGCGAAUAAUA------------------AUUAAUCACCAUGCUCACCAUGAAAUCAAUAGACCCCAUUGAUUUCCACUUCUGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGU .((((..((.......------------------.....................(((((((((.......)))))))))...((((((((......)).))))))....))..)))).. ( -22.10) >DroEre_CAF1 67476 106 - 1 AUUAUAGCAGAUUAUUAAUGU----GUGAAAUGUAUUAAUCACAAUGCUCGCCAUGAAAUCAAUAGACCCCAUUGGUUUCCAUUUCUGCGUGCAAAAACAGCAGAAAAAA---------- .....((((....(((((((.----.(...)..))))))).....))))......(((((((((.......)))))))))..(((((((((......)).)))))))...---------- ( -22.00) >DroYak_CAF1 69718 119 - 1 AUAAUAGCGGGUUAUUAAUGUACAUAUGUAAUGUGAAAAUCGCCAUGCACACCAUGAAAUCAAUAGACCCGAGUCAUUGCCAUUUCUGCGUACAAAAACAGCAGAAAAAA-GAAAUGCGU .......((((((.......(((((.....)))))........((((.....)))).........)))))).(.((((....(((((((((......)).)))))))...-..))))).. ( -23.40) >consensus UGCAUAGCGGAUAAUA__________________AUUAAUCACCAUGCUCACCAUGAAAUCAAUAGACCCCAUUGAUUUCCACUUCUGCGUGCAAAAACAGCAGAAAAAACGAAAUGCGU .......................................................(((((((((.......)))))))))...((((((((......)).)))))).............. (-10.42 = -11.26 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:07 2006