| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,247,382 – 19,247,480 |
| Length | 98 |
| Max. P | 0.976228 |

| Location | 19,247,382 – 19,247,480 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -32.08 |
| Energy contribution | -31.70 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
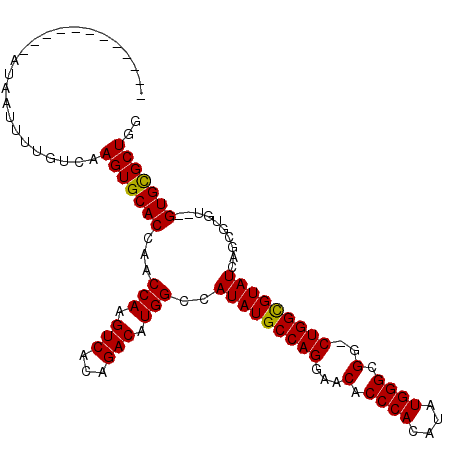
>2L_DroMel_CAF1 19247382 98 + 22407834 ------------AUAAUUUUGUCAAGUGCACCAACCAAGUCACAGACAUGGCCAUAUGCCAGGAACACCCACAUAUGGGCGG-CUGGUGUAUCAGCGUGUGUGUGUGCUGG ------------.............((((((((.((..(((...)))..(.(((((((...((.....)).))))))).)))-.))))))))(((((........))))). ( -31.40) >DroSec_CAF1 67294 96 + 1 ------------AUAAUUUUGUUAAGUGCACCAACCAAGUCACAGACAUGGCCAUAUGCCAGGAACACCCACAUAUGGGCGG-CUGGCGUAUCAGCGUGU--GUGCGCUGG ------------............(((((((((.(((.(((...))).)))..(((((((((...(.((((....)))).).-))))))))).....)).--))))))).. ( -33.80) >DroSim_CAF1 68631 96 + 1 ------------AUAAUUUUGUUGAGUGCACCAACCAAGUCACAGACAUGGCCAUAUGCCAGGAACACCCACAUAUGGGCGG-CUGGCGUAUCAGCGUGU--GUGCGCUGG ------------............(((((((((.(((.(((...))).)))..(((((((((...(.((((....)))).).-))))))))).....)).--))))))).. ( -33.90) >DroYak_CAF1 68733 107 + 1 AUCAUUUUUAUA-UAAUUUUUGCAAGUGCAC-AACCAAGUCACAGACAUGGCCAUAUGCCAGGAACACCCACAUAUGGGCGGACUGGCGUAUCAGCGUGU--GUGCGCUGG ............-...........(((((((-(.(((.(((...))).)))..(((((((((...(.((((....)))).)..))))))))).......)--))))))).. ( -35.80) >consensus ____________AUAAUUUUGUCAAGUGCACCAACCAAGUCACAGACAUGGCCAUAUGCCAGGAACACCCACAUAUGGGCGG_CUGGCGUAUCAGCGUGU__GUGCGCUGG ........................(((((((...(((.(((...))).)))..(((((((((...(.((((....)))).)..)))))))))..........))))))).. (-32.08 = -31.70 + -0.38)

| Location | 19,247,382 – 19,247,480 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -29.86 |
| Energy contribution | -29.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
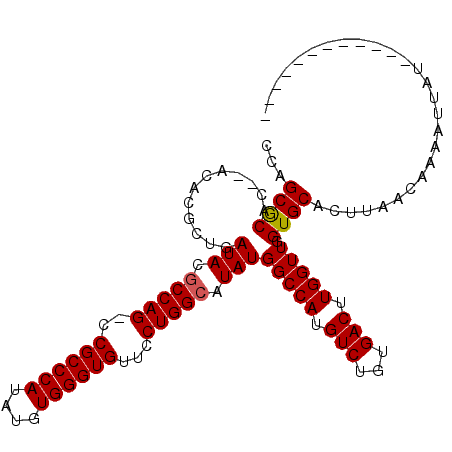
>2L_DroMel_CAF1 19247382 98 - 22407834 CCAGCACACACACACGCUGAUACACCAG-CCGCCCAUAUGUGGGUGUUCCUGGCAUAUGGCCAUGUCUGUGACUUGGUUGGUGCACUUGACAAAAUUAU------------ .((((..........))))...((((((-(((.((((((((.((.....)).)))))))).)..(((...)))..))))))))................------------ ( -31.20) >DroSec_CAF1 67294 96 - 1 CCAGCGCAC--ACACGCUGAUACGCCAG-CCGCCCAUAUGUGGGUGUUCCUGGCAUAUGGCCAUGUCUGUGACUUGGUUGGUGCACUUAACAAAAUUAU------------ .(((((...--...)))))...((((((-(((.((((((((.((.....)).)))))))).)..(((...)))..))))))))................------------ ( -33.90) >DroSim_CAF1 68631 96 - 1 CCAGCGCAC--ACACGCUGAUACGCCAG-CCGCCCAUAUGUGGGUGUUCCUGGCAUAUGGCCAUGUCUGUGACUUGGUUGGUGCACUCAACAAAAUUAU------------ .(((((...--...)))))...((((((-(((.((((((((.((.....)).)))))))).)..(((...)))..))))))))................------------ ( -33.90) >DroYak_CAF1 68733 107 - 1 CCAGCGCAC--ACACGCUGAUACGCCAGUCCGCCCAUAUGUGGGUGUUCCUGGCAUAUGGCCAUGUCUGUGACUUGGUU-GUGCACUUGCAAAAAUUA-UAUAAAAAUGAU .(((((...--...)))))....(((((..((((((....))))))...)))))..(..((((.(((...))).)))).-.)((....))........-............ ( -35.20) >consensus CCAGCGCAC__ACACGCUGAUACGCCAG_CCGCCCAUAUGUGGGUGUUCCUGGCAUAUGGCCAUGUCUGUGACUUGGUUGGUGCACUUAACAAAAUUAU____________ ...((((............(((.(((((..((((((....))))))...))))).)))(((((.(((...))).))))).))))........................... (-29.86 = -29.93 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:02 2006