
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,246,853 – 19,246,943 |
| Length | 90 |
| Max. P | 0.534099 |

| Location | 19,246,853 – 19,246,943 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -17.65 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
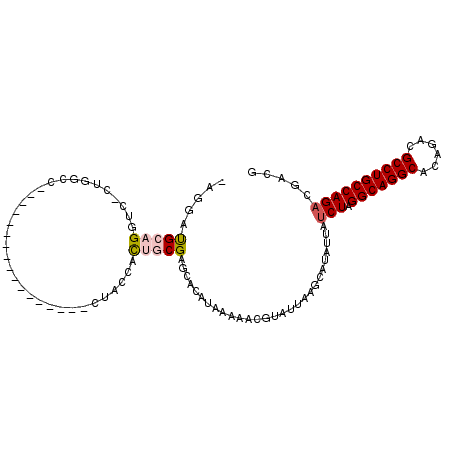
>2L_DroMel_CAF1 19246853 90 + 22407834 AAGGAUGCAGGUC-CUGGCC-----------------CUACCACUGCGAGCACAUAAAAACGUAUUAAGCAUAUUAUCUAGGCAGGCACAGACGCCUGCCAGACGACG ....((((.(((.-...)))-----------------.......((((............))))....)))).(..(((.(((((((......))))))))))..).. ( -25.00) >DroPse_CAF1 75156 107 + 1 CAGGUCGCAGGAC-GAGGCUCCAGAAUGGAAAUGGAACCAGAAUGCCGGGCCCAUAAAAACGUAUUAAGCAUAUUAUCUAGGCAGGCACAGACGCCUGCCAGACAACG ...(((...(((.-.(.(((((.....))).((((..((........))..)))).............))...)..))).(((((((......))))))).))).... ( -32.80) >DroSec_CAF1 66753 90 + 1 -AGGAUGCAGGUCCCUGGCC-----------------CUACCACUGCGAGCACAUAAAAACGUAUUAAGCAUAUUAUCUAGGCAGGCACAGACGCCUGCCAGACGACG -...((((.(((.....)))-----------------.......((((............))))....)))).(..(((.(((((((......))))))))))..).. ( -25.00) >DroEre_CAF1 66100 89 + 1 -AGGAUGCAGGUC-CUGGCC-----------------CUACCACUGCGAGCACAUAAAAACGUAUUAAGCAUAUUAUCUAGGCAGGCACAGACGCCUGCCAGCCGACG -(((((....)))-))(((.-----------------.......(((....((........)).....))).........(((((((......))))))).))).... ( -25.80) >DroYak_CAF1 68213 89 + 1 -CGGAUGCAGGUC-CUGGCC-----------------CUACCACUGCGAGCACAUAAAAACGUAUUAAGCAUAUUAUCUAGGCAGGCACAGACGCCUGCCAGACGACG -((.((((.(((.-...)))-----------------.......((((............))))....))))....(((.(((((((......))))))))))))... ( -25.40) >DroPer_CAF1 78289 107 + 1 CAGGUCGCAGGAC-GAGGUUCCAGAAUGGAAAUGGAACCAGAAUGCCGGGCCCAUAAAAACGUAUUAAGCAUAUUAUCUAGGCAGGCACAGACGCCUGCCAGACAACG ..(((((((...(-..(((((((.........))))))).)..)))..))))........................(((.(((((((......))))))))))..... ( -33.80) >consensus _AGGAUGCAGGUC_CUGGCC_________________CUACCACUGCGAGCACAUAAAAACGUAUUAAGCAUAUUAUCUAGGCAGGCACAGACGCCUGCCAGACGACG .....(((((.................................)))))............................(((.(((((((......))))))))))..... (-17.65 = -18.04 + 0.39)

| Location | 19,246,853 – 19,246,943 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -20.01 |
| Energy contribution | -19.60 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
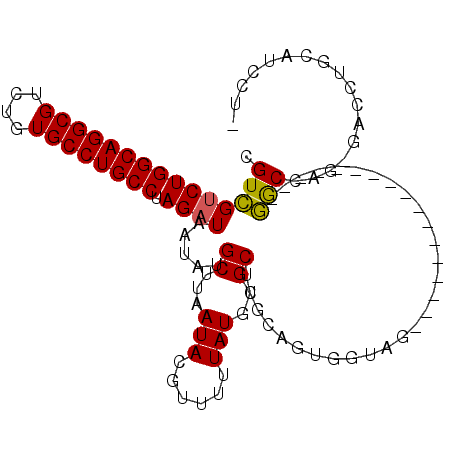
>2L_DroMel_CAF1 19246853 90 - 22407834 CGUCGUCUGGCAGGCGUCUGUGCCUGCCUAGAUAAUAUGCUUAAUACGUUUUUAUGUGCUCGCAGUGGUAG-----------------GGCCAG-GACCUGCAUCCUU .((..(((((((((((....)))))))).)))..)).(((....(((((....)))))...)))...((((-----------------(.....-..)))))...... ( -29.60) >DroPse_CAF1 75156 107 - 1 CGUUGUCUGGCAGGCGUCUGUGCCUGCCUAGAUAAUAUGCUUAAUACGUUUUUAUGGGCCCGGCAUUCUGGUUCCAUUUCCAUUCUGGAGCCUC-GUCCUGCGACCUG .(((((((((((((((....)))))))).)))))))..((((...........((((..((((....))))..))))..........)))).((-(.....))).... ( -37.20) >DroSec_CAF1 66753 90 - 1 CGUCGUCUGGCAGGCGUCUGUGCCUGCCUAGAUAAUAUGCUUAAUACGUUUUUAUGUGCUCGCAGUGGUAG-----------------GGCCAGGGACCUGCAUCCU- .((..(((((((((((....)))))))).)))..)).(((....(((((....)))))...)))...((((-----------------(.(....).))))).....- ( -30.00) >DroEre_CAF1 66100 89 - 1 CGUCGGCUGGCAGGCGUCUGUGCCUGCCUAGAUAAUAUGCUUAAUACGUUUUUAUGUGCUCGCAGUGGUAG-----------------GGCCAG-GACCUGCAUCCU- ....((...(((((..((((..((((((.........(((....(((((....)))))...)))..)))))-----------------)..)))-).)))))..)).- ( -28.30) >DroYak_CAF1 68213 89 - 1 CGUCGUCUGGCAGGCGUCUGUGCCUGCCUAGAUAAUAUGCUUAAUACGUUUUUAUGUGCUCGCAGUGGUAG-----------------GGCCAG-GACCUGCAUCCG- .((..(((((((((((....)))))))).)))..)).(((....(((((....)))))...)))...((((-----------------(.....-..))))).....- ( -29.60) >DroPer_CAF1 78289 107 - 1 CGUUGUCUGGCAGGCGUCUGUGCCUGCCUAGAUAAUAUGCUUAAUACGUUUUUAUGGGCCCGGCAUUCUGGUUCCAUUUCCAUUCUGGAACCUC-GUCCUGCGACCUG .(((((((((((((((....)))))))).))))))).(((.....(((.....((((..((((....))))..))))((((.....))))...)-))...)))..... ( -37.10) >consensus CGUCGUCUGGCAGGCGUCUGUGCCUGCCUAGAUAAUAUGCUUAAUACGUUUUUAUGUGCUCGCAGUGGUAG_________________GGCCAG_GACCUGCAUCCU_ .(((((((((((((((....)))))))).)))).....((...(((......)))..)).............................)))................. (-20.01 = -19.60 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:01 2006