
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,236,743 – 19,236,885 |
| Length | 142 |
| Max. P | 0.697781 |

| Location | 19,236,743 – 19,236,861 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.79 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
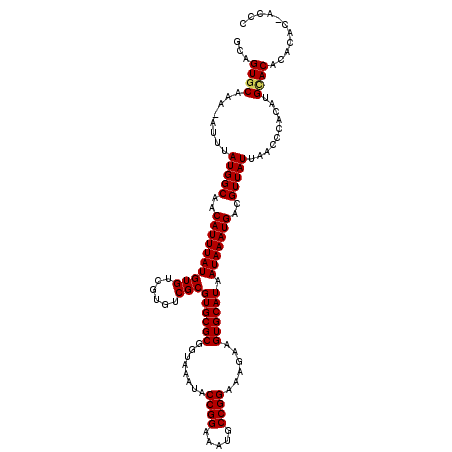
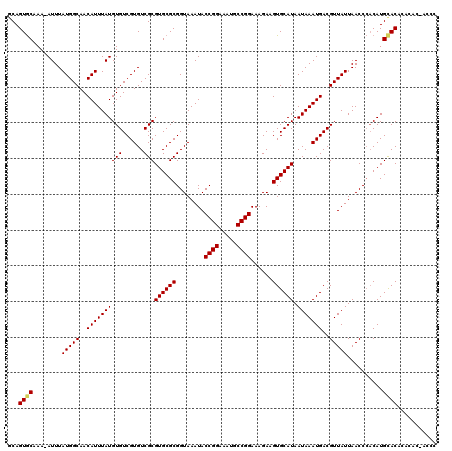
>2L_DroMel_CAF1 19236743 118 + 22407834 GCAGUGCAAA-AUUUAUGGCAACAUUUAUGUGUCGUGUCGCGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACUCACAC-ACAC (.((((((..-((..(((....)))..))..(((((.....((((((........((((.....))))......)))))).....)))))..............)))))))....-.... ( -30.24) >DroVir_CAF1 69103 119 + 1 GCAGUGCUAA-AUUUAUGGCAACAUUUAUGUGUCGUGUCGCGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGUACACACCCCACCG ....(((((.-.....)))))........(((..((((.(((((.(.(((.((((((((.....))))......((.(((.....)))))..)))).)))).)))))..))))..))).. ( -28.70) >DroGri_CAF1 72871 120 + 1 CCAGUGCUAAAAUUUAUGGCAACAUUUAUGUGUCGUGUCGCGUGCGCGGUAAAUACCGGAAAUACCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCGUCAUGCACACAAGCCAACC ................((((.((((....)))).((((.(((((.(((((.((((((((.....))))......((.(((.....)))))..)))).))))))))))))))..))))... ( -34.80) >DroSec_CAF1 56608 116 + 1 GCAGUGCAAA-AUUUAUGGCAACAUUUAUGUGUCGUGUCGCGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACUCACAC-A--C (.((((((..-((..(((....)))..))..(((((.....((((((........((((.....))))......)))))).....)))))..............)))))))....-.--. ( -30.24) >DroMoj_CAF1 81076 119 + 1 GCAGUGCUAA-AUUUAUGGCAACAUUUAUGUGUCGUGUCGCGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACACAGCCCACCC ((..(((((.-.....)))))((((....)))).((((.(((((.(.(((.((((((((.....))))......((.(((.....)))))..)))).)))).))))))))).))...... ( -30.70) >DroAna_CAF1 107134 112 + 1 GCAGUACAAA-AUUUAUGGCAACAUUUAUGUGUCGUGUCGCGUGCGCGGUAAAUACCGGAAAUACCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCAC-------ACAC ...(.(((..-....(((....))).....))))((((.(((((.(.(((.((((((((.....))))......((.(((.....)))))..)))).)))).)))))))-------)).. ( -29.20) >consensus GCAGUGCAAA_AUUUAUGGCAACAUUUAUGUGUCGUGUCGCGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACACACAC_ACCC ...((((........(((((..((((((((((......)))((((((........((((.....))))......)))))).)))))))..)))))..........))))........... (-26.74 = -26.79 + 0.06)

| Location | 19,236,782 – 19,236,885 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.04 |
| Mean single sequence MFE | -20.89 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.05 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
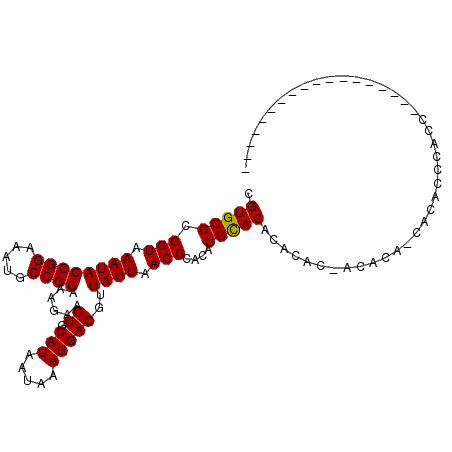
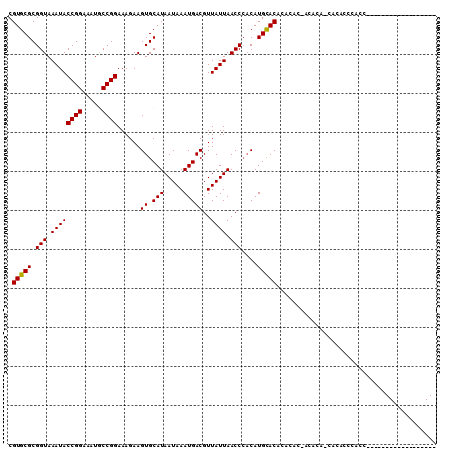
>2L_DroMel_CAF1 19236782 103 + 22407834 CGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACUCACAC-ACACA-CACACUCACGCGGGAGUAGA-----ACA .(((.(.(((.....((((.....))))...((.(((((((((((......)))))).......))))))))).)-.).))-)..((((......))))...-----... ( -21.41) >DroVir_CAF1 69142 92 + 1 CGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGUACACACCCCACCGGUCCCCCCGUCC------------------ .(((.(.(((.....((((.....))))......(((((((((((......)))))).......)))))..)))))))(((.....)))...------------------ ( -18.61) >DroGri_CAF1 72911 92 + 1 CGUGCGCGGUAAAUACCGGAAAUACCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCGUCAUGCACACAAGCCAACCAUCUUCCCACCC------------------ .(((((((.(.....((((.....)))).....).))).......((((((........)))))))))).......................------------------ ( -21.40) >DroEre_CAF1 56296 103 + 1 CGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACUCUCAC-ACACA-CACUACCAACCGGGAGUAGA-----ACA .(((.(.(.......((((.....))))...((((((((((((((......)))))).......)))))).)).)-.).))-)(((.((....)).)))...-----... ( -21.61) >DroMoj_CAF1 81115 92 + 1 CGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACACAGCCCACCCACCACGCCCACG------------------ ((((.(((.......((((.....))))......(((((((((((......)))))).......)))))...............))).))))------------------ ( -22.01) >DroAna_CAF1 107173 102 + 1 CGUGCGCGGUAAAUACCGGAAAUACCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCAC-------ACACA-UCCAUUCGGCGGAGAGUAGAACAGAACA .(((.(((.......((((.....))))......(((..((((((......))))))...))).))).)-------))...-((.((((......)))).))........ ( -20.30) >consensus CGUGCGCGGUAAAUACCGGAAAUGCCGGAAAGAAGUGCAUAAUAAAUGACGUUAUUAACCCACAUGCACACACAC_ACACA_CACACCCACC__________________ .(((((.(((.((((((((.....))))......((.(((.....)))))..)))).)))....)))))......................................... (-16.19 = -16.05 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:51 2006