
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,234,499 – 19,234,612 |
| Length | 113 |
| Max. P | 0.529214 |

| Location | 19,234,499 – 19,234,612 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -9.79 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19234499 113 - 22407834 UGGCCAUAAUGAAAAUGUAAUUUCAUUUCUGGCCAAAAGACCCAUCACUUAAAACACUCCUCCACUACUCGGCCCACUCGGCCAGCUCCUCAUCAUCGUAUCUGCCUGCGUGC ((((((.(((((((......)))))))..))))))...................(((.............((((.....))))..............(((......)))))). ( -24.30) >DroPse_CAF1 61332 92 - 1 UCGAAAUAAUGAAAAUGUAAUUUCAUUUCGGACCAAAAUACCCAUCACUCAGG-CACUGU---GGGU---GCCACACCACACCACCCC-------------CUACCC-CCUAC (((((...((((((......)))))))))))...................(((-....((---((((---(......))).))))..)-------------))....-..... ( -15.20) >DroSec_CAF1 54349 111 - 1 UGGCCAUAAUGAAAAUGUAAUUUCAUUUCUGGCCAAAACACCCAUUACUUAAAACACGCCCCCACUC--UGCCCCACUCGGCCAGCUCCUCAUCAUCGUAUCUGCCUGCGUGC ((((((.(((((((......)))))))..))))))...................(((((........--.(((......)))..((.................))..))))). ( -24.03) >DroSim_CAF1 56092 111 - 1 UGGCCAUAAUGAAAAUGUAAUUUCAUUUCUGGCCAAAAGACCCAUCACUUAAAACACGCCCCCACUC--UGCCCCUCUCGGCCAGCUCCUCAUCAUCGUAUCUGCCUGUGUGC ((((((.(((((((......)))))))..))))))...................(((((........--.(((......)))..((.................))..))))). ( -22.13) >DroEre_CAF1 53941 99 - 1 UGGCCGUAAUGAAAAUGUAAUUUCAUUUCUGCCCAAAAGACCCAUCACUGAAAACACUCC---AUUC--AGC-----CCGGCCAGCUCCUCAUCAUCGUAUCUG----CGUGC ((((((.(((((((......)))))))....................(((((........---.)))--)).-----.)))))).........((.(((....)----)))). ( -17.20) >DroYak_CAF1 55898 99 - 1 UGGCCAUAAUGAAAAUGUAAUUUCAUUUCUGGCCAAAAGACCCAUCACUUAAAACACUCC---GUUC--AGC-----CCGGCCAGCUCCUCAUCGUCGUAUCUG----CGUGC ((((((.(((((((......)))))))..))))))........................(---(..(--((.-----.((((.(........).))))...)))----..)). ( -18.80) >consensus UGGCCAUAAUGAAAAUGUAAUUUCAUUUCUGGCCAAAAGACCCAUCACUUAAAACACUCC___ACUC__AGCCCCACCCGGCCAGCUCCUCAUCAUCGUAUCUGCCU_CGUGC ((((((.(((((((......)))))))..)))))).............................................................................. ( -9.79 = -10.52 + 0.72)

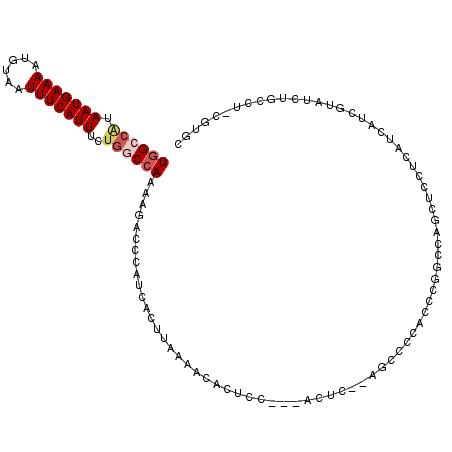
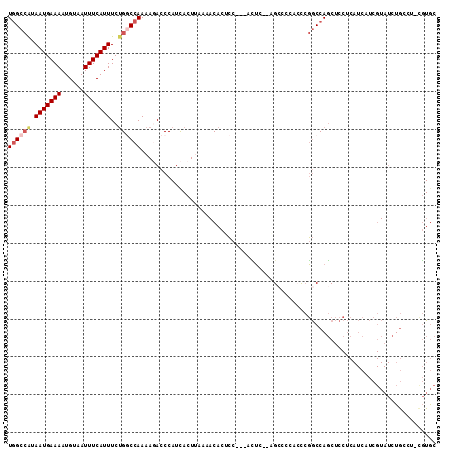
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:49 2006