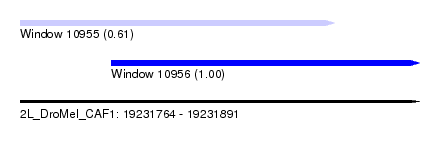
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,231,764 – 19,231,891 |
| Length | 127 |
| Max. P | 0.998181 |

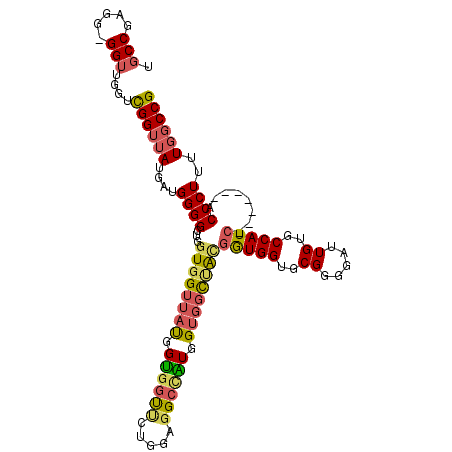
| Location | 19,231,764 – 19,231,864 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.27 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -34.30 |
| Energy contribution | -33.76 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.605001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19231764 100 + 22407834 AAGGUCCUUUCACCGAAGGUGGUGGCUUCUGCCGAGGGGGUUGGUUGGUUAUGAUGGGGCUGGUGGUUAUGGCUAU------GGCUGUGGUGGCCAUGGUGGUGCG ..((((((..((((...))))((((((...(((((.....))))).))))))...))))))....((.((.(((((------((((.....))))))))).)))). ( -34.90) >DroSec_CAF1 51633 105 + 1 AAGGUCCUUUCGCCGAAGGUGGUGGCUUCUGCCGAGG-GGUUGGUCGGUUAUGAUGGGGCUGGUGGUUACGGUGGUUCUGGAGGCCAUGGUGGCUAUGGUGGUGCG ..((((((.(((((((......((((....))))...-......)))))...)).)))))).((((((((.((((((.....)))))).))))))))......... ( -37.16) >DroSim_CAF1 53382 105 + 1 AAGGUCCUUUCGCCGAAGGUGGUGGCUUCUGCCGAGG-GGUUGGUCGGUUAUGAUGGGGCUGGUGGUUAUGGUGGUUCUGGAGGCCAUGGUGGCUACGGUGGUGCG ..((((((.(((((((......((((....))))...-......)))))...)).)))))).((((((((.((((((.....)))))).))))))))......... ( -37.66) >consensus AAGGUCCUUUCGCCGAAGGUGGUGGCUUCUGCCGAGG_GGUUGGUCGGUUAUGAUGGGGCUGGUGGUUAUGGUGGUUCUGGAGGCCAUGGUGGCUAUGGUGGUGCG ..((((((.(((((((......((((....))))..........)))))...)).)))))).((((((((.((((((.....)))))).))))))))......... (-34.30 = -33.76 + -0.54)

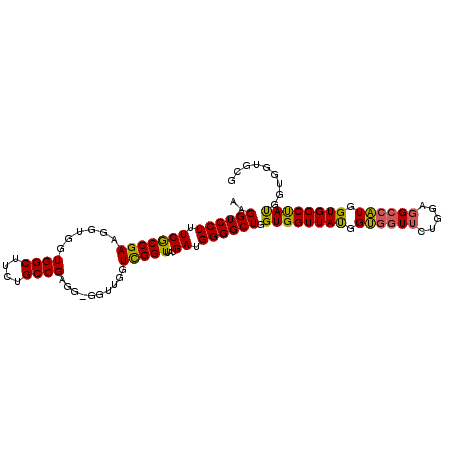
| Location | 19,231,793 – 19,231,891 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -30.23 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19231793 98 + 22407834 UGCCGAGGGGGUUGGUUGGUUAUGAUGGGGCUGGUGGUUAUGGCUAU------GGCUGUGGUGGCCAUGGUGGUGCGGGGAAUGUGCCAUC-------ACCCUUUUGGCCG .(((((((((((....(((((((.((..(((((.((((....)))))------)))))).))))))).(((((..((.....))..)))))-------))))))))))).. ( -43.70) >DroSec_CAF1 51662 103 + 1 UGCCGAGG-GGUUGGUCGGUUAUGAUGGGGCUGGUGGUUACGGUGGUUCUGGAGGCCAUGGUGGCUAUGGUGGUGCGGGGAUUGUGCCAUC-------ACCCUUUUGGCCG .(((((((-(((....(((((.......)))))((((((((.((((((.....)))))).))))))))(((((..((.....))..)))))-------)))).)))))).. ( -42.80) >DroSim_CAF1 53411 103 + 1 UGCCGAGG-GGUUGGUCGGUUAUGAUGGGGCUGGUGGUUAUGGUGGUUCUGGAGGCCAUGGUGGCUACGGUGGUGCGGGGAUUGUGCCAUC-------ACCCUUUUGGCCG .(((((((-(((....(((((.......)))))((((((((.((((((.....)))))).))))))))(((((..((.....))..)))))-------)))).)))))).. ( -43.60) >DroEre_CAF1 50909 103 + 1 CGCCGAGG-GG----UUGGUUAUGAUGGGGCUGUUGGUUAUGGUGGCCAUGGUGGCUGUGG---UGGCUAUGGUGCGAGGAUUGUGCCAUCACCCAUCACCCUUUUUUCCG ....((((-((----(.......((((((.....(((((((.(..(((.....)))..).)---))))))(((..(((...)))..)))...)))))))))))))...... ( -39.41) >consensus UGCCGAGG_GGUUGGUCGGUUAUGAUGGGGCUGGUGGUUAUGGUGGUUCUGGAGGCCAUGGUGGCUACGGUGGUGCGGGGAUUGUGCCAUC_______ACCCUUUUGGCCG .(((.....)))....((((((....((((...((((((((.((((((.....)))))).))))))))(((((..((.....))..)))))........))))..)))))) (-30.23 = -31.10 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:47 2006