| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,229,293 – 19,229,383 |
| Length | 90 |
| Max. P | 0.993146 |

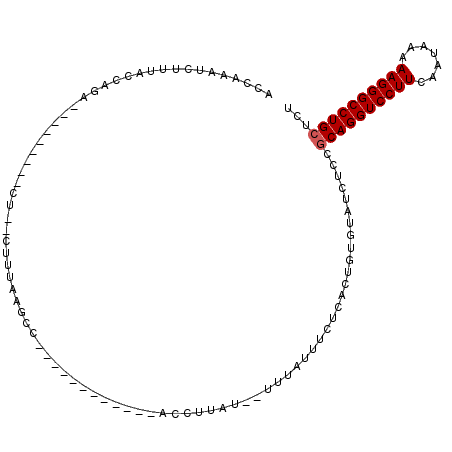
| Location | 19,229,293 – 19,229,383 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 64.54 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -13.66 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19229293 90 + 22407834 GGAGCAGGCCCUUUUUAUUGAAGGACCUGCGGAGAUACACAGUAAGAAAUAAC--UUUAUUU------------UAUUUAAAGAGAG---------UCUGGUAAAGAUUUGGU ...(((((.(((((.....))))).))))).((((((...(((.(....).))--).)))))------------).........(((---------(((.....))))))... ( -21.50) >DroSec_CAF1 49152 90 + 1 AGAGCAGGCCCUUUUUAUUGAAGGACCUGUGGAGAUAUACAGUGAGAAAUAAA--AUGAAGU------------GGCUGAAAGAGAG---------UCUGGCAAAGAUUUGGU ...(((((.(((((.....))))).))))).........((((.(........--......)------------.)))).....(((---------(((.....))))))... ( -19.04) >DroSim_CAF1 50909 76 + 1 GGAGCAGGCCCUUUUUAUUGAAGGACCUGCGGAGAUACACAGUGAGAAAUAAA--AUAAAGU------------GGCUUAA-----------------------AGAUGUGGU ...(((((.(((((.....))))).))))).......((((.((((..((...--.....))------------..)))).-----------------------...)))).. ( -21.90) >DroEre_CAF1 48591 88 + 1 AGAGCAGGCCCUU-UUAUUGAAGGACCUGCGGAGUUACACAGUGAGAAAUGCACGCUAAGAA------------------------GGCAGUAAGGACUGGUCAUGUUUUGUC .(((((((.((((-(....))))).)))))..((((..((.((((....).)))(((.....------------------------))).))...))))..)).......... ( -23.20) >DroYak_CAF1 50775 111 + 1 AGAGCAGGCCCUU-UUAUUGAAGGACCUGCGGAGUUACACGGUAAGAAAUGCACAGUAAGAAGGACCUUAAUUUGUCUGCAAGUAAGGCAGUUAGGG-UGGUAAUGAUUUGUC ...(((((.((((-(....))))).)))))...(((((((.(((.....)))...)).......(((((((((.((((.......))))))))))))-).)))))........ ( -31.90) >consensus AGAGCAGGCCCUUUUUAUUGAAGGACCUGCGGAGAUACACAGUGAGAAAUAAA__AUAAAAU____________GGCUGAAAG__AG_________UCUGGUAAAGAUUUGGU ...(((((.(((((.....))))).)))))................................................................................... (-13.66 = -13.90 + 0.24)

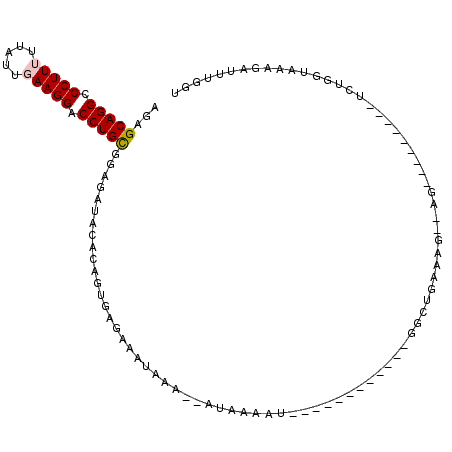
| Location | 19,229,293 – 19,229,383 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.54 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19229293 90 - 22407834 ACCAAAUCUUUACCAGA---------CUCUCUUUAAAUA------------AAAUAAA--GUUAUUUCUUACUGUGUAUCUCCGCAGGUCCUUCAAUAAAAAGGGCCUGCUCC ..........((((((.---------....(((((....------------...))))--)..........))).))).....((((((((((.......))))))))))... ( -19.36) >DroSec_CAF1 49152 90 - 1 ACCAAAUCUUUGCCAGA---------CUCUCUUUCAGCC------------ACUUCAU--UUUAUUUCUCACUGUAUAUCUCCACAGGUCCUUCAAUAAAAAGGGCCUGCUCU .................---------.............------------.......--........................(((((((((.......))))))))).... ( -13.00) >DroSim_CAF1 50909 76 - 1 ACCACAUCU-----------------------UUAAGCC------------ACUUUAU--UUUAUUUCUCACUGUGUAUCUCCGCAGGUCCUUCAAUAAAAAGGGCCUGCUCC .........-----------------------......(------------((.....--.............))).......((((((((((.......))))))))))... ( -17.07) >DroEre_CAF1 48591 88 - 1 GACAAAACAUGACCAGUCCUUACUGCC------------------------UUCUUAGCGUGCAUUUCUCACUGUGUAACUCCGCAGGUCCUUCAAUAA-AAGGGCCUGCUCU ...........((((((......((((------------------------........).)))......)))).))......((((((((((......-))))))))))... ( -22.90) >DroYak_CAF1 50775 111 - 1 GACAAAUCAUUACCA-CCCUAACUGCCUUACUUGCAGACAAAUUAAGGUCCUUCUUACUGUGCAUUUCUUACCGUGUAACUCCGCAGGUCCUUCAAUAA-AAGGGCCUGCUCU .............((-(.....((((.......))))(((...(((((....))))).)))............))).......((((((((((......-))))))))))... ( -25.40) >consensus ACCAAAUCUUUACCAGA_________CU__CUUUAAGCC____________ACCUUAU__UUUAUUUCUCACUGUGUAUCUCCGCAGGUCCUUCAAUAAAAAGGGCCUGCUCU ...................................................................................((((((((((.......))))))))))... (-15.64 = -15.84 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:44 2006