| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,217,418 – 19,217,521 |
| Length | 103 |
| Max. P | 0.965907 |

| Location | 19,217,418 – 19,217,521 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.55 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
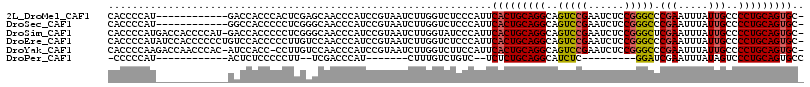
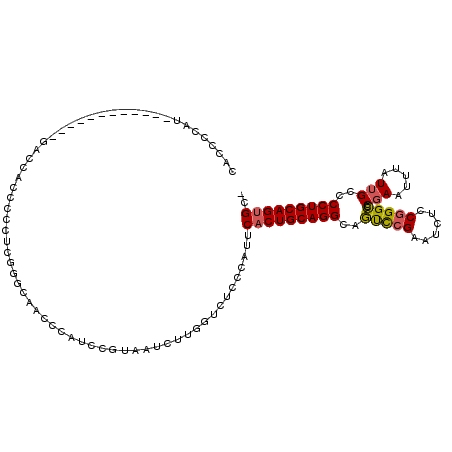
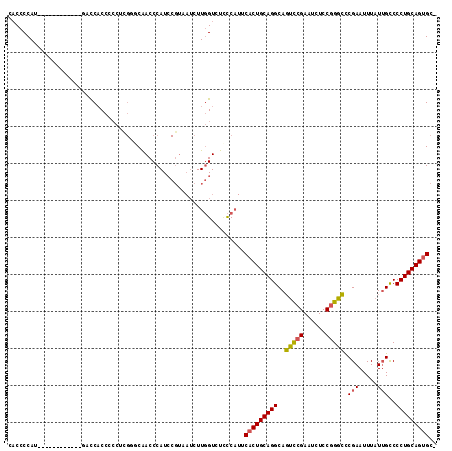
>2L_DroMel_CAF1 19217418 103 - 22407834 CACCCCAU------------GACCACCCACUCGAGCAACCCAUCCGUAAUCUUGGUCUCCCAUUCACUGCAGGCAGUCCGAAUCUCCGGGCCCGAAUUUAUUGCCCCUGCAGUGC- ........------------(((((......((...........))......))))).......(((((((((..(((((......))))).(((.....)))..))))))))).- ( -26.70) >DroSec_CAF1 37316 103 - 1 CACCCCAU------------GGCCACCCCCUCGGGCAACCCAUCCGUAAUCUUGGUCUCCCAUUCACUGCAGGCAGUCCGAAUCUCCGGGCCCGAAUUUAUUGCCCCUGCAGUGC- ........------------(((((......((((.......))))......))))).......(((((((((..(((((......))))).(((.....)))..))))))))).- ( -30.20) >DroSim_CAF1 39093 114 - 1 CACCCCAUGACCACCCCAU-GACCACCCCCUCGGGCAACCCAUCCGUAAUCUUGGUAUCCCAUUCACUGCAGGCAGUCCGAAUCUCCGGGCUCGAAUUUAUUGCCCCUGCAGUGC- .....((((.......)))-)((((......((((.......))))......))))........(((((((((.((((((......))))))(((.....)))..))))))))).- ( -30.90) >DroEre_CAF1 36837 115 - 1 CACCCCAUAUCCACCCCCCUGUCCACCCCCUUGUCCAACCCAUCCGUAAUCUUGGUCUCCCAUUCACUGCAGGCAGUCCGAAUCUCCGGGCCCGAAUUUAUUGCCCCUGCAGUGC- ................................(.((((.............)))).).......(((((((((..(((((......))))).(((.....)))..))))))))).- ( -22.52) >DroYak_CAF1 38723 113 - 1 CACCCCAAGACCAACCCAC-AUCCACC-CCUUGUCCAACCCAUCCGUAAUCUUGGUCUUCCAUUCACUGCAGGCAGUCCGAAUCUCCGGGCCCGAAUUUAUUGCCCCUGCAGUGC- ......((((((((...((-(......-...)))...((......))....)))))))).....(((((((((..(((((......))))).(((.....)))..))))))))).- ( -27.30) >DroPer_CAF1 46197 83 - 1 -CCCCCAU------------ACUCUCCCCCUU--UCGACCCAU-------CUUUGUCUGUC--UCUCUGCAGGCAUCUC---------GGAUCGAAUUUAUAGUCCCUGCAGUGCC -.......------------...........(--((((.((..-------...(((((((.--.....)))))))....---------)).))))).................... ( -15.10) >consensus CACCCCAU____________GACCACCCCCUCGGGCAACCCAUCCGUAAUCUUGGUCUCCCAUUCACUGCAGGCAGUCCGAAUCUCCGGGCCCGAAUUUAUUGCCCCUGCAGUGC_ ................................................................(((((((((..(((((......))))).(((.....)))..))))))))).. (-18.56 = -18.55 + -0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:40 2006