
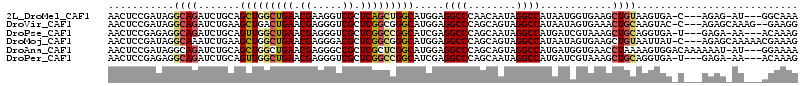
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,203,531 – 19,203,638 |
| Length | 107 |
| Max. P | 0.832561 |

| Location | 19,203,531 – 19,203,638 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.609994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
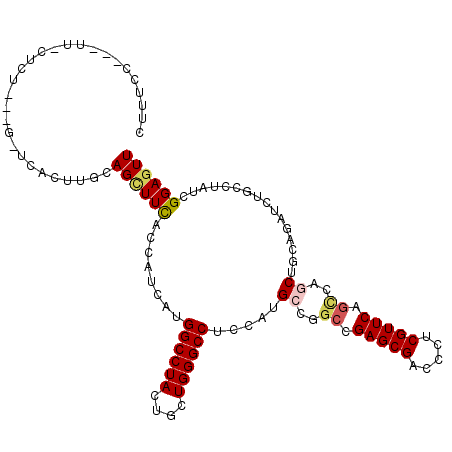
>2L_DroMel_CAF1 19203531 107 + 22407834 UUUGCC---AU-CUCU---G-UCACUUACAGCUUCACCAUUAUGGCCUAUUGUUGGGCCUCCAUGCCAGCUGAGCGACCUUCGUUCAGCCAGCUGCAGAUCUGCCUAUCGGAGUU ......---..-((((---(-........((((....(((...((((((....))))))...)))...((((((((.....)))))))).))))(((....)))....))))).. ( -33.60) >DroVir_CAF1 36411 109 + 1 CCUUC--CUUUGCUCU---G-GUACUUGCAGUUUCACUAUUAUGGCCUACUGCUGGGCCUCCAUGCCCGCCGAGCGACCCUCGUUCAGUCAGCUUCAGAUCUGCCUAUCGGAGUU .....--....(((((---(-(((...((((..((........((((((....)))))).....((..((.(((((.....))))).))..))....)).)))).))))))))). ( -34.50) >DroPse_CAF1 27917 107 + 1 CUUUGU---UU-UCUC---A-UCACCUGCAGCUUUACGAUCAUGGCCUAUUGCUGGGCCUCGAUGCCGGCCGAGCGACCCUCGUUCAGCCAACUGCAGAUCUGCCUCUCGGAGUU ......---..-....---.-....((((((.......(((..((((((....))))))..)))...(((.(((((.....))))).)))..)))))).((((.....))))... ( -34.70) >DroMoj_CAF1 42297 111 + 1 CUUUCGUUUUUGCUCU---G-AUAAUUACAGCUUCACUAUUAUGGCCUACUGCUGGGCCUCCAUGCCCGCCGAGCGUCCCUCGUUCAGCCAGCUUCAGAUUUGCCUAUCGGAGUU ...........(((((---(-(((..................((((.....((.((((......)))))).(((((.....))))).))))((.........)).))))))))). ( -30.70) >DroAna_CAF1 72620 111 + 1 UUUUCC---AU-AUUUUUUGUCCACUUUUAGGUUCACCAUCAUGGCCUACUGCUGGGCCUCCAUGCCGAGCGAGCGGCCCUCGUUCAGCCAGCUGCAGAUCUGCCUAUCGGAGUU ......---..-.......(((((.(..(((((.((......)))))))..).)))))((((..((.(((((((.....))))))).)).....(((....))).....)))).. ( -29.90) >DroPer_CAF1 30803 107 + 1 CUUUGU---UU-UCUC---A-UCACCUGCAGCUUUACGAUCAUGGCCUAUUGCUGGGCCUCGAUGCCGGCCGAGCGACCCUCGUUCAGCCAACUGCAGAUCUGCCUCUCGGAGUU ......---..-....---.-....((((((.......(((..((((((....))))))..)))...(((.(((((.....))))).)))..)))))).((((.....))))... ( -34.70) >consensus CUUUCC___UU_CUCU___G_UCACUUGCAGCUUCACCAUCAUGGCCUACUGCUGGGCCUCCAUGCCGGCCGAGCGACCCUCGUUCAGCCAGCUGCAGAUCUGCCUAUCGGAGUU .............................((((((........((((((....)))))).....((..((.(((((.....))))).))..))................)))))) (-18.74 = -19.13 + 0.39)

| Location | 19,203,531 – 19,203,638 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.15 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
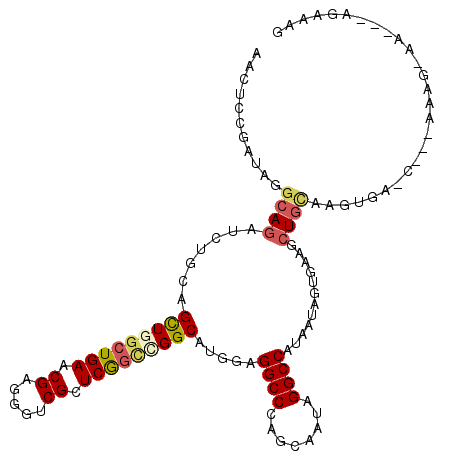
>2L_DroMel_CAF1 19203531 107 - 22407834 AACUCCGAUAGGCAGAUCUGCAGCUGGCUGAACGAAGGUCGCUCAGCUGGCAUGGAGGCCCAACAAUAGGCCAUAAUGGUGAAGCUGUAAGUGA-C---AGAG-AU---GGCAAA ..(((((....(((....))).((..(((((.((.....)).)))))..)).)))))(((.........(((.....)))....((((.....)-)---))..-..---)))... ( -37.00) >DroVir_CAF1 36411 109 - 1 AACUCCGAUAGGCAGAUCUGAAGCUGACUGAACGAGGGUCGCUCGGCGGGCAUGGAGGCCCAGCAGUAGGCCAUAAUAGUGAAACUGCAAGUAC-C---AGAGCAAAG--GAAGG ...(((........(((((....(.....).....)))))((((((.((((......)))).(((((....(((....)))..))))).....)-)---.))))...)--))... ( -30.20) >DroPse_CAF1 27917 107 - 1 AACUCCGAGAGGCAGAUCUGCAGUUGGCUGAACGAGGGUCGCUCGGCCGGCAUCGAGGCCCAGCAAUAGGCCAUGAUCGUAAAGCUGCAGGUGA-U---GAGA-AA---ACAAAG ..((((....)).))((((((((((((((((.((.....)).)))))).((((((.((((.(....).)))).)))).))..))))))))))..-.---....-..---...... ( -41.70) >DroMoj_CAF1 42297 111 - 1 AACUCCGAUAGGCAAAUCUGAAGCUGGCUGAACGAGGGACGCUCGGCGGGCAUGGAGGCCCAGCAGUAGGCCAUAAUAGUGAAGCUGUAAUUAU-C---AGAGCAAAAACGAAAG ..(((.(((((((....(((..(((.(((((.((.....)).))))).))).(((....))).)))...)))...((((.....))))...)))-)---.)))......(....) ( -32.40) >DroAna_CAF1 72620 111 - 1 AACUCCGAUAGGCAGAUCUGCAGCUGGCUGAACGAGGGCCGCUCGCUCGGCAUGGAGGCCCAGCAGUAGGCCAUGAUGGUGAACCUAAAAGUGGACAAAAAAU-AU---GGAAAA ...((((...(((....((((.....(((((.((((.....)))).))))).(((....)))))))...))).....((....))..................-.)---)))... ( -32.90) >DroPer_CAF1 30803 107 - 1 AACUCCGAGAGGCAGAUCUGCAGUUGGCUGAACGAGGGUCGCUCGGCCGGCAUCGAGGCCCAGCAAUAGGCCAUGAUCGUAAAGCUGCAGGUGA-U---GAGA-AA---ACAAAG ..((((....)).))((((((((((((((((.((.....)).)))))).((((((.((((.(....).)))).)))).))..))))))))))..-.---....-..---...... ( -41.70) >consensus AACUCCGAUAGGCAGAUCUGCAGCUGGCUGAACGAGGGUCGCUCGGCCGGCAUGGAGGCCCAGCAAUAGGCCAUAAUAGUGAAGCUGCAAGUGA_C___AAAG_AA___AGAAAG ...........((((.......(((((((((.((.....)).))))))))).....((((........))))............))))........................... (-24.70 = -25.15 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:37 2006