| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
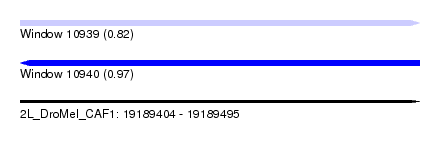
| Location | 19,189,404 – 19,189,495 |
| Length | 91 |
| Max. P | 0.974938 |

| Location | 19,189,404 – 19,189,495 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -12.42 |
| Energy contribution | -12.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
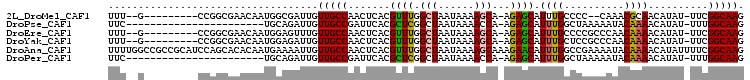
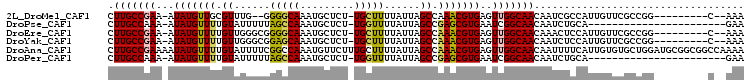
>2L_DroMel_CAF1 19189404 91 + 22407834 UUU--G---------CCGGCGAACAAUGGCGAUUGUUGCCAACUCACGUUUGGCUAAUAAAAGCA-AGAGCAUUUGCCCC--CAAACGCAACAUAU-UUCGGCAAG .((--(---------(((((.((((((....)))))))))......(((((((....((((.((.-...)).))))...)--))))))........-...))))). ( -27.20) >DroPse_CAF1 13985 81 + 1 UUC-----------------------UGCAGAUUGUUGCCGAUUCACGCUCGGCUAAUAAAACCA-AGAGCAUUUGGCUAAAAAUACAAAACAUAU-UUUGGCAAG (((-----------------------(.....((((((((((.......))))).))))).....-))))......(((((((............)-))))))... ( -15.20) >DroEre_CAF1 9579 93 + 1 UUU--G---------CCGGCGAACAAUGGAGUUUGUUGCCAACUCACGUUUGGCUAAUAAAAGCA-AGAGCAUUUGCCCCGCCCAACAAAACAUAU-UUCGGCAAG .((--(---------(((((.(((((......)))))))).......(((.(((...((((.((.-...)).))))....))).))).........-...))))). ( -24.70) >DroYak_CAF1 10513 93 + 1 UUU--G---------CCGGCGAACAAUGGAGAUUGUUGCCAACUCACGUUUGGCUAAUAAAAGCA-AGAGCAUUUGCUCCGCCCAACAAAACAUAU-UUCGGCAAG .((--(---------(((((.((((((....))))))))).......((((.(((......))).-.((((....)))).........))))....-...))))). ( -26.20) >DroAna_CAF1 59799 106 + 1 UUUUGGCCGCCGCAUCCAGCACACAAUGAAAAUUGUUGCCAACUCACGUUUGGCUAAUAAAAGCAAAGAACAUUUGGCCGAAAAUACAAAACAUAUUUUCGGCAAG ..(((((....((.....))..(((((....))))).))))).....((((.(((......)))...)))).....((((((((((.......))))))))))... ( -29.20) >DroPer_CAF1 16824 81 + 1 UUC-----------------------UGCAGAUUGUUGCCGAUUCACGCUCGGCUAAUAAAACCA-AGAGCAUUUGGCUAAAAAUACAAAACAUAU-UUUGGCAAG (((-----------------------(.....((((((((((.......))))).))))).....-))))......(((((((............)-))))))... ( -15.20) >consensus UUU__G_________CCGGCGAACAAUGGAGAUUGUUGCCAACUCACGUUUGGCUAAUAAAAGCA_AGAGCAUUUGCCCCAAAAAACAAAACAUAU_UUCGGCAAG ...................................(((((.......((((.(((......)))...)))).((((..........))))..........))))). (-12.42 = -12.20 + -0.22)
| Location | 19,189,404 – 19,189,495 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.05 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
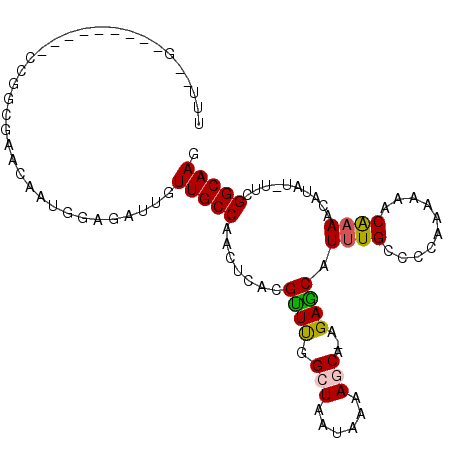
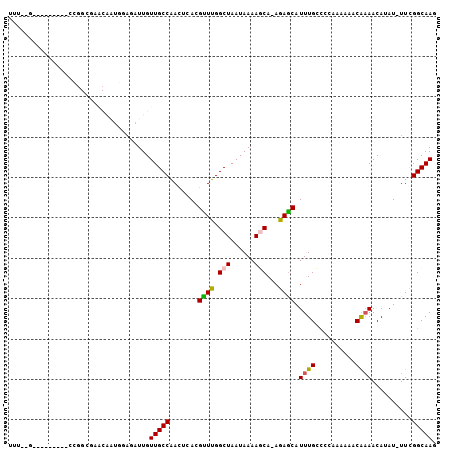
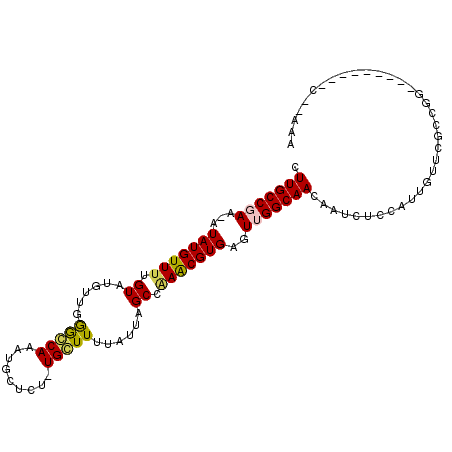
>2L_DroMel_CAF1 19189404 91 - 22407834 CUUGCCGAA-AUAUGUUGCGUUUG--GGGGCAAAUGCUCU-UGCUUUUAUUAGCCAAACGUGAGUUGGCAACAAUCGCCAUUGUUCGCCGG---------C--AAA .(((((...-.....(..((((((--(((((....)))))-.(((......)))))))))..)...(((((((((....)))))).)))))---------)--)). ( -32.80) >DroPse_CAF1 13985 81 - 1 CUUGCCAAA-AUAUGUUUUGUAUUUUUAGCCAAAUGCUCU-UGGUUUUAUUAGCCGAGCGUGAAUCGGCAACAAUCUGCA-----------------------GAA .((((.(((-((((.....)))))))..(((..((((((.-.((((.....)))))))))).....)))........)))-----------------------).. ( -19.00) >DroEre_CAF1 9579 93 - 1 CUUGCCGAA-AUAUGUUUUGUUGGGCGGGGCAAAUGCUCU-UGCUUUUAUUAGCCAAACGUGAGUUGGCAACAAACUCCAUUGUUCGCCGG---------C--AAA .(((((...-.(((((((.((((((((((((....)).))-)))).....)))).)))))))....((((((((......))))).)))))---------)--)). ( -30.20) >DroYak_CAF1 10513 93 - 1 CUUGCCGAA-AUAUGUUUUGUUGGGCGGAGCAAAUGCUCU-UGCUUUUAUUAGCCAAACGUGAGUUGGCAACAAUCUCCAUUGUUCGCCGG---------C--AAA .(((((...-.(((((((.((((((((((((....)))).-)))).....)))).)))))))....(((((((((....)))))).)))))---------)--)). ( -31.60) >DroAna_CAF1 59799 106 - 1 CUUGCCGAAAAUAUGUUUUGUAUUUUCGGCCAAAUGUUCUUUGCUUUUAUUAGCCAAACGUGAGUUGGCAACAAUUUUCAUUGUGUGCUGGAUGCGGCGGCCAAAA ...(((((((((((.....)))))))))))...(((((....(((......)))..)))))...(((((.(((((....))))).(((((....)))))))))).. ( -30.70) >DroPer_CAF1 16824 81 - 1 CUUGCCAAA-AUAUGUUUUGUAUUUUUAGCCAAAUGCUCU-UGGUUUUAUUAGCCGAGCGUGAAUCGGCAACAAUCUGCA-----------------------GAA .((((.(((-((((.....)))))))..(((..((((((.-.((((.....)))))))))).....)))........)))-----------------------).. ( -19.00) >consensus CUUGCCGAA_AUAUGUUUUGUAUGUUGGGCCAAAUGCUCU_UGCUUUUAUUAGCCAAACGUGAGUUGGCAACAAUCUCCAUUGUUCGCCGG_________C__AAA .(((((((...(((((((.((......(((((.........)))))......)).)))))))..)))))))................................... (-15.47 = -15.05 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:32 2006