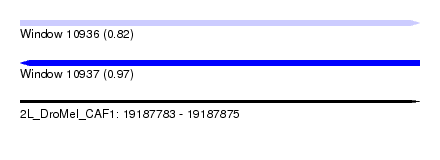
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,187,783 – 19,187,875 |
| Length | 92 |
| Max. P | 0.972184 |

| Location | 19,187,783 – 19,187,875 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 71.50 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -11.57 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19187783 92 + 22407834 AAAGGAAAGGGUGAAGCGGCCCAAGUUGUGCCACAUCCAU-GUGGCAUCAUAAAACACAUGCAAAGUGUUUGUUUUG---------------UAGGCAGUGUGUUCUG ...((((.((((......))))..((.((((((((....)-))))))).))...((((.(((((((......)))))---------------))....)))).)))). ( -25.90) >DroSec_CAF1 7818 75 + 1 AAGGGAAAGGGUGAAGCGGCCCAAGUUGUGCCACAUCCAU-GUGGCAUCAUAAAACACAUGCAAAGUGUUUGUUU--------------------------------G ........((((......))))..((.((((((((....)-))))))).)).((((((.......))))))....--------------------------------. ( -21.80) >DroSim_CAF1 9080 75 + 1 AAGGGAAAGGGUGAAGCGGCCCAAGUUGUGCCACAUCCAU-GUGGCAUCAUAAAACACAUGCAAAGUGUUUGCUU--------------------------------G ........((((......))))((((.((((((((....)-)))))))....((((((.......))))))))))--------------------------------. ( -22.60) >DroEre_CAF1 7960 107 + 1 AAAGGAAAGGGUGAAGCGGCCCAAGUUGGGCCACAUCCAU-GUGGGAUCAUAAAACACAUGCAAAGUGUUUGUUCGGUGAGCGGCAGAGGGGUGGGCAGUGUUUUCUG ..((((((.(.((....(((((.....))))).(((((.(-((.(..((((.((((((.......)))))).....)))).).)))...)))))..)).).)))))). ( -37.80) >DroYak_CAF1 8888 105 + 1 G-AGAAAAGGGUGAAGCGGCCCAAGUUGGGCCACAUCCAU-GUGGAAUCAUAAAACGCAUGCAAAGUGUUUGCUUGG-GAGUGGCAAAGGGGUGGGCAGUGGGUUCUU .-....(((..(...((.(((((..((..(((((.(((((-(((...........)))))((((.....))))...)-)))))))...))..))))).))..)..))) ( -34.20) >DroAna_CAF1 58151 86 + 1 AAGGGAAAGGGU-----GGCCAAGGUGGGGACAAAUCCACCGAGGAAUCGUAAAACACAUGCAAAGUGUUUGUUUGUUU-------------UGGG---U-GAUUGUC .........(((-----.(((..((((((......))))))..(((((....((((((.......))))))....))))-------------).))---)-.)))... ( -21.50) >consensus AAAGGAAAGGGUGAAGCGGCCCAAGUUGGGCCACAUCCAU_GUGGCAUCAUAAAACACAUGCAAAGUGUUUGUUUGG_______________UGGG___U___UUCUG ........(((((....(((.........))).)))))..............((((((.......))))))..................................... (-11.57 = -11.77 + 0.19)

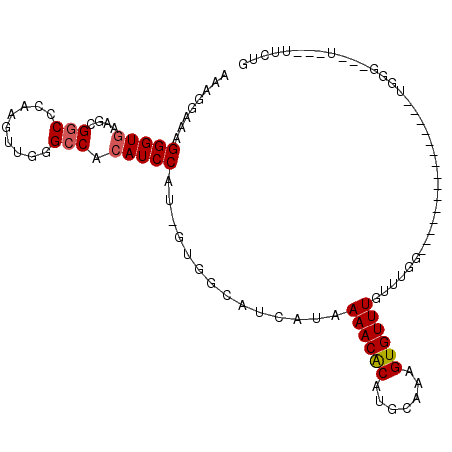
| Location | 19,187,783 – 19,187,875 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.50 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -9.32 |
| Energy contribution | -10.48 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19187783 92 - 22407834 CAGAACACACUGCCUA---------------CAAAACAAACACUUUGCAUGUGUUUUAUGAUGCCAC-AUGGAUGUGGCACAACUUGGGCCGCUUCACCCUUUCCUUU .((((((((.(((...---------------...............))))))))))).((.((((((-(....)))))))))....(((........)))........ ( -22.67) >DroSec_CAF1 7818 75 - 1 C--------------------------------AAACAAACACUUUGCAUGUGUUUUAUGAUGCCAC-AUGGAUGUGGCACAACUUGGGCCGCUUCACCCUUUCCCUU .--------------------------------....((((((.......))))))..((.((((((-(....)))))))))....(((........)))........ ( -18.60) >DroSim_CAF1 9080 75 - 1 C--------------------------------AAGCAAACACUUUGCAUGUGUUUUAUGAUGCCAC-AUGGAUGUGGCACAACUUGGGCCGCUUCACCCUUUCCCUU .--------------------------------((((((((((.......))))))..((.((((((-(....))))))))).........))))............. ( -19.00) >DroEre_CAF1 7960 107 - 1 CAGAAAACACUGCCCACCCCUCUGCCGCUCACCGAACAAACACUUUGCAUGUGUUUUAUGAUCCCAC-AUGGAUGUGGCCCAACUUGGGCCGCUUCACCCUUUCCUUU ((.((((((((((............((.....))............))).))))))).))((((...-..))))(((((((.....)))))))............... ( -25.75) >DroYak_CAF1 8888 105 - 1 AAGAACCCACUGCCCACCCCUUUGCCACUC-CCAAGCAAACACUUUGCAUGCGUUUUAUGAUUCCAC-AUGGAUGUGGCCCAACUUGGGCCGCUUCACCCUUUUCU-C .(((((.((.(((.......(((((.....-....)))))......))))).))))).(((.(((..-..))).(((((((.....))))))).))).........-. ( -25.12) >DroAna_CAF1 58151 86 - 1 GACAAUC-A---CCCA-------------AAACAAACAAACACUUUGCAUGUGUUUUACGAUUCCUCGGUGGAUUUGUCCCCACCUUGGCC-----ACCCUUUCCCUU .......-.---.(((-------------(.......((((((.......))))))...........(((((........)))))))))..-----............ ( -15.40) >consensus CAGAA___A___CCCA_______________CCAAACAAACACUUUGCAUGUGUUUUAUGAUGCCAC_AUGGAUGUGGCACAACUUGGGCCGCUUCACCCUUUCCCUU .....................................((((((.......)))))).............((((.(((((.........)))))))))........... ( -9.32 = -10.48 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:30 2006