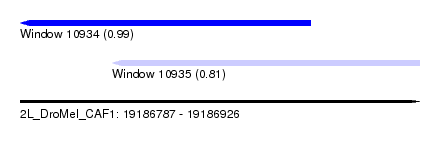
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,186,787 – 19,186,926 |
| Length | 139 |
| Max. P | 0.989954 |

| Location | 19,186,787 – 19,186,888 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.35 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19186787 101 - 22407834 -----------ACAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGAACGAACGCACACAAAGUGGCAAAUAAUUGCCCAGUUAAUUGAAAAGCCAAAAGGCAG-------- -----------..........((((.(((((((((((....))))))).............((.(((...))))).......)))).)))).........(((....)))..-------- ( -21.00) >DroSec_CAF1 6794 112 - 1 CAAACGAGGAGCCAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGACGAACACACACAAAGUGGCAAAUAAUUGCCCAGUUAAUUGAAAAGCCAAAAGGCAG-------- ....(((..(((.......(((((((....(((((((....))))))))))))))...............(.(((((....)))))).)))..)))....(((....)))..-------- ( -26.50) >DroSim_CAF1 8053 112 - 1 CAAACGAGGAGCCAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGACGAACGCACACAAAGUGGCAAAUAAUUGCCCAGUUAAUUGAAAAGCCAAAAGGCAG-------- ....(((..(((.......(((((((....(((((((....))))))))))))))...............(.(((((....)))))).)))..)))....(((....)))..-------- ( -26.50) >DroEre_CAF1 7005 111 - 1 CAAACGAGA-----AAAUCCCGGCUUGGCAUAAAUCAAAUCUGAUUUAAAGUCGGAC----GCACACAAAGUGGCAAAUAAUUGCCAAGUUAAUUGAAAAGCCAAAAGGCGACUGGAGUU .........-----...(((.(((((((((.........((((((.....)))))).----((.(((...))))).......))))))))).........(((....)))....)))... ( -28.60) >DroYak_CAF1 7969 111 - 1 CAAACAAGG-----AAAUUCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGAC----GCACACAAAGUGGCAAAUAAUUGCCAAGUUAAUUGAAAAGCCAAAAGGCGACUUAAGUU ....(((((-----.....))((((((((((((((((....))))))).........----((.(((...))))).......)))))))))..)))....(((....))).......... ( -27.40) >consensus CAAACGAGG___CAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGACGAACGCACACAAAGUGGCAAAUAAUUGCCCAGUUAAUUGAAAAGCCAAAAGGCAG________ ...................(((((((....(((((((....)))))))))))))).................(((((....)))))..............(((....))).......... (-21.72 = -21.92 + 0.20)



| Location | 19,186,819 – 19,186,926 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.26 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -20.30 |
| Energy contribution | -22.30 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19186819 107 - 22407834 UAAAUAAUCCCAGCAAGUCGCA-UUCGUUUGCCCAGGAG------------ACAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGAACGAACGCACACAAAGUGGCAAAUA ............((.....((.-((((((((((..(((.------------......))).)))..(((.(((((((....)))))))..)))))))))).)).(((...)))))..... ( -25.60) >DroSec_CAF1 6826 119 - 1 UAAAUAAUCCCAGCUAGUCGCA-UUCGUUUGCCCAGGAGCCAAACGAGGAGCCAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGACGAACACACACAAAGUGGCAAAUA ...................((.-...(((((.((....(((((.((.(((.......)))))..))))).(((((((....))))))).....)).))))).(((.....)))))..... ( -29.60) >DroSim_CAF1 8085 119 - 1 UAAAUAAUCCCAGCUAGUCGCA-UUCGUUUGCCCAGGAGCCAAACGAGGAGCCAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGACGAACGCACACAAAGUGGCAAAUA ............((.....((.-(((((((((......(((((.((.(((.......)))))..))))).(((((((....)))))))..).)))))))).)).(((...)))))..... ( -30.70) >DroEre_CAF1 7045 111 - 1 UAAAUAAUCCCAGGAAGUCGCAAUUCGUUUGCCCAAGAGCCAAACGAGA-----AAAUCCCGGCUUGGCAUAAAUCAAAUCUGAUUUAAAGUCGGAC----GCACACAAAGUGGCAAAUA .......((....)).(((((..(((((((((......).)))))))).-----.....(((((((....(((((((....))))))))))))))..----.........)))))..... ( -27.80) >DroYak_CAF1 8009 111 - 1 UAAAUAAUCCCAGCAAGUCGCAUUUCGUUUGCCCAGGAGCCAAACAAGG-----AAAUUCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGAC----GCACACAAAGUGGCAAAUA .......(((..(((((.((.....)))))))...)))((((.......-----....((((((((....(((((((....))))))))))))))).----..........))))..... ( -27.95) >consensus UAAAUAAUCCCAGCAAGUCGCA_UUCGUUUGCCCAGGAGCCAAACGAGG___CAAAAUCCCGGCUUGGCAUAAAUCAAAUUUGAUUUAAAGUCGGACGAACGCACACAAAGUGGCAAAUA ................(((((..((((((((.(.....).))))))))...........(((((((....(((((((....))))))))))))))...............)))))..... (-20.30 = -22.30 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:28 2006