| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,186,654 – 19,186,747 |
| Length | 93 |
| Max. P | 0.827653 |

| Location | 19,186,654 – 19,186,747 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
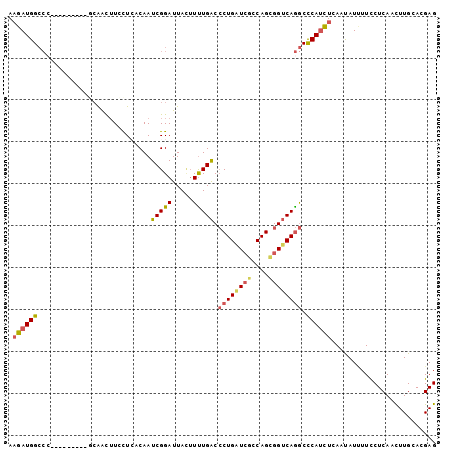
>2L_DroMel_CAF1 19186654 93 + 22407834 AAGAUGGCCC---------CCAACUUGCUAACAAUCGGAUUACUUUUGACCCUGAUCGCCAGCGGUCAGGCCCAUCUCAAUAUUUUCCUCAACUUGCACGAG .((((((...---------...............(((((.....))))).((((((((....)))))))).))))))......................... ( -21.10) >DroGri_CAF1 20627 102 + 1 AUAAUGGAGAAUAACUUUAAAGGCAUCUUCACAAUCGGAUUAUGUUUGGCGCUGCUCAGCAGCGGACAAACCCAUUUGAAUAUAUUUCUCAAUUUGCACGAG ......(((((....((((((((.((((........))))...(((((.((((((...))))))..))))))).)))))).....)))))............ ( -22.40) >DroSim_CAF1 7920 93 + 1 AAGAUGGCCC---------CCAACUUGCUCACAAUCGGAUUACUUUUGACCCUGAUCGCCAGCGGUCAGGCCCAUCUCAAUAUUUUCCUCAACUUGCACGAG .((((((...---------...............(((((.....))))).((((((((....)))))))).))))))......................... ( -21.10) >DroEre_CAF1 6872 93 + 1 AAGAUGGCCC---------GCAACUUCCUCACAAUCGGAUUACUUUUGACCCUGAUCGCCAGCGGUCAGGCUCAUCUAAAUAUUUUCCUCAACUUGCAUGAG .((((((((.---------......(((........))).......((((((((.....))).)))))))).)))))..........((((.......)))) ( -24.00) >DroYak_CAF1 7836 93 + 1 AAGAUGGCCC---------GCAACUUCCUCUCAAUCGGAUUACUUUUGACCCUGAUCGCCAGCGGUCAGGCUCAUCUAAACAUUUUCCUCAACUUGCACGAG .((((((((.---------......(((........))).......((((((((.....))).)))))))).)))))......................... ( -22.70) >DroAna_CAF1 56954 102 + 1 AAGAUGGCCAACAGCCGGAGCAAUUUGCUGACAAUCGGAUUAAUUCUGACCCUGGUAGCCAGUGGUCAGGCCCACCUCAACAUCUUCCUCAACUUGCACGAG ((((((((.....)))((.((((((..(((.....)))...))))((((((((((...)))).))))))))))........)))))................ ( -27.50) >consensus AAGAUGGCCC_________GCAACUUCCUCACAAUCGGAUUACUUUUGACCCUGAUCGCCAGCGGUCAGGCCCAUCUCAAUAUUUUCCUCAACUUGCACGAG .((((((...........................(((((.....))))).((((((((....)))))))).))))))......................... (-15.48 = -15.65 + 0.17)

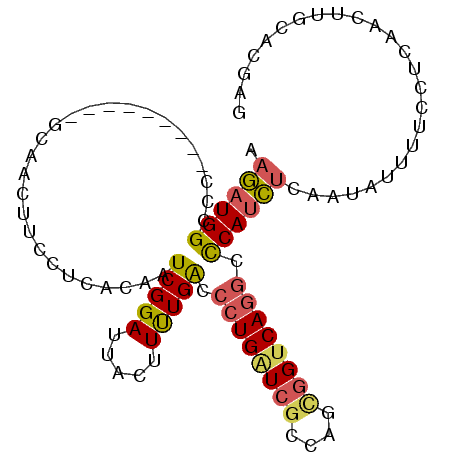
| Location | 19,186,654 – 19,186,747 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.05 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
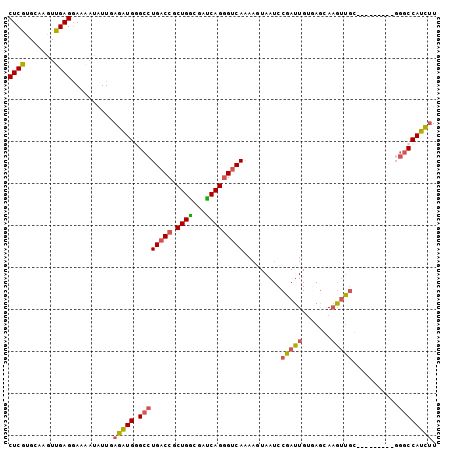
>2L_DroMel_CAF1 19186654 93 - 22407834 CUCGUGCAAGUUGAGGAAAAUAUUGAGAUGGGCCUGACCGCUGGCGAUCAGGGUCAAAAGUAAUCCGAUUGUUAGCAAGUUGG---------GGGCCAUCUU ((((.......)))).........(((((((.((..((.((((((((((.((((........))))))))))))))..))..)---------.).))))))) ( -34.50) >DroGri_CAF1 20627 102 - 1 CUCGUGCAAAUUGAGAAAUAUAUUCAAAUGGGUUUGUCCGCUGCUGAGCAGCGCCAAACAUAAUCCGAUUGUGAAGAUGCCUUUAAAGUUAUUCUCCAUUAU ((((.......))))...........((((((((((..((((((...)))))).))))).......((((.(((((....))))).)))).....))))).. ( -23.80) >DroSim_CAF1 7920 93 - 1 CUCGUGCAAGUUGAGGAAAAUAUUGAGAUGGGCCUGACCGCUGGCGAUCAGGGUCAAAAGUAAUCCGAUUGUGAGCAAGUUGG---------GGGCCAUCUU ((((.......)))).........(((((((.((..((.(((.((((((.((((........)))))))))).)))..))..)---------.).))))))) ( -33.40) >DroEre_CAF1 6872 93 - 1 CUCAUGCAAGUUGAGGAAAAUAUUUAGAUGAGCCUGACCGCUGGCGAUCAGGGUCAAAAGUAAUCCGAUUGUGAGGAAGUUGC---------GGGCCAUCUU ((((.......))))..........(((((.((((((((.(((.....)))))))....(((((((........)))..))))---------))))))))). ( -28.50) >DroYak_CAF1 7836 93 - 1 CUCGUGCAAGUUGAGGAAAAUGUUUAGAUGAGCCUGACCGCUGGCGAUCAGGGUCAAAAGUAAUCCGAUUGAGAGGAAGUUGC---------GGGCCAUCUU ((((.......))))..........(((((.((((((((.(((.....)))))))....(((((((........)))..))))---------))))))))). ( -27.80) >DroAna_CAF1 56954 102 - 1 CUCGUGCAAGUUGAGGAAGAUGUUGAGGUGGGCCUGACCACUGGCUACCAGGGUCAGAAUUAAUCCGAUUGUCAGCAAAUUGCUCCGGCUGUUGGCCAUCUU ((((.......)))).........(((((((..((((((.((((...)))))))))).......(((((.((((((.....)))..))).)))))))))))) ( -34.70) >consensus CUCGUGCAAGUUGAGGAAAAUAUUGAGAUGGGCCUGACCGCUGGCGAUCAGGGUCAAAAGUAAUCCGAUUGUGAGCAAGUUGC_________GGGCCAUCUU ((((.......))))..........(((((.((((((((.((((...))))))))).........(((((.......)))))...........)))))))). (-18.93 = -19.05 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:26 2006