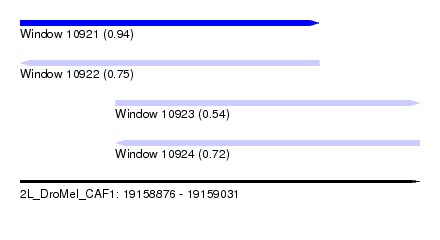
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,158,876 – 19,159,031 |
| Length | 155 |
| Max. P | 0.937983 |

| Location | 19,158,876 – 19,158,992 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -21.84 |
| Energy contribution | -24.40 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19158876 116 + 22407834 AAAUCUAU---UUUUCUAUUAAAACUUGGGACUAUUUUGCAUUUUGACAUACGCACCGCACAAUAUUAUGCCCACCACUGUUGGCAGUGGGUGUGUGUGUCAUGGUAAUUGCCUUUGUU ........---...........(((..(((.(....((((....((((((((((((((((........)))....(((((....))))))))))))))))))..))))..))))..))) ( -37.60) >DroSim_CAF1 39407 116 + 1 AAAUCUUU---UUUUCUAUUAAAACGUGUGCCUACUUUGCAUUUUGACAUACGCAGCGCUCAAUAUUAUGCCCACCACUGUUGGCAGUGGGUGUGUGUGUCAUGGUAAUUGCCUUUGUU ........---...........((((...((.....((((....((((((((((((.((..........)).).((((((....)))))).)))))))))))..))))..))...)))) ( -30.30) >DroYak_CAF1 37722 111 + 1 AACUCUGUUCCUUUUUUAUUAGAACAUGUACAUAUUUUGCA--------UGAGCACCGCCCAAUAUUGGGCCCACCACUGUUGUCAGUUGGUGUGUGUGUCAUGGUAAUUGCCUUUGUU .....(((((...........))))).............((--------(((((((.(((((....))))).((((((((....))).))))).)))).)))))............... ( -28.20) >consensus AAAUCUAU___UUUUCUAUUAAAACAUGUGCCUAUUUUGCAUUUUGACAUACGCACCGCACAAUAUUAUGCCCACCACUGUUGGCAGUGGGUGUGUGUGUCAUGGUAAUUGCCUUUGUU ....................................((((....((((((((((((((((........)))....(((((....))))))))))))))))))..))))........... (-21.84 = -24.40 + 2.56)

| Location | 19,158,876 – 19,158,992 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -18.19 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19158876 116 - 22407834 AACAAAGGCAAUUACCAUGACACACACACCCACUGCCAACAGUGGUGGGCAUAAUAUUGUGCGGUGCGUAUGUCAAAAUGCAAAAUAGUCCCAAGUUUUAAUAGAAAA---AUAGAUUU .......(((.((....(((((.((.((((..(..(((....)))..)(((((....))))))))).)).))))))).)))...........................---........ ( -26.20) >DroSim_CAF1 39407 116 - 1 AACAAAGGCAAUUACCAUGACACACACACCCACUGCCAACAGUGGUGGGCAUAAUAUUGAGCGCUGCGUAUGUCAAAAUGCAAAGUAGGCACACGUUUUAAUAGAAAA---AAAGAUUU ....(((((........(((((.((.(((((((..(.....)..)))))((......)).....)).)).)))))...(((.......)))...))))).........---........ ( -22.30) >DroYak_CAF1 37722 111 - 1 AACAAAGGCAAUUACCAUGACACACACACCAACUGACAACAGUGGUGGGCCCAAUAUUGGGCGGUGCUCA--------UGCAAAAUAUGUACAUGUUCUAAUAAAAAAGGAACAGAGUU .......((((((..((((((((...(((((.(((....)))))))).(((((....))))).))).)))--------))...))).)))...((((((.........))))))..... ( -31.30) >consensus AACAAAGGCAAUUACCAUGACACACACACCCACUGCCAACAGUGGUGGGCAUAAUAUUGAGCGGUGCGUAUGUCAAAAUGCAAAAUAGGCACAAGUUUUAAUAGAAAA___AAAGAUUU .......(((.....((((...(((.((((.((((....)))))))).(((((....))))).)))..))))......)))...................................... (-18.19 = -18.87 + 0.67)


| Location | 19,158,913 – 19,159,031 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -27.21 |
| Energy contribution | -30.10 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19158913 118 + 22407834 AUUUUGACAUACGCACCGCACAAUAUUAUGCCCACCACUGUUGGCAGUGGGUGUGUGUGUCAUGGUAAUUGCCUUUGUUUUUGCUGCCGCUAUUGCUGCUUUUGGUACCAUUGAACUC ....((((((((((((((((........)))....(((((....))))))))))))))))))((((....(((...((....)).((.((....)).))....)))))))........ ( -38.80) >DroSim_CAF1 39444 118 + 1 AUUUUGACAUACGCAGCGCUCAAUAUUAUGCCCACCACUGUUGGCAGUGGGUGUGUGUGUCAUGGUAAUUGCCUUUGUUUUUGCUGCCGCUAUUGCUGCUUUUGGUACCAUUGAACUC ....((((((((((((.((..........)).).((((((....)))))).)))))))))))((((....(((...((....)).((.((....)).))....)))))))........ ( -34.80) >DroYak_CAF1 37762 110 + 1 A--------UGAGCACCGCCCAAUAUUGGGCCCACCACUGUUGUCAGUUGGUGUGUGUGUCAUGGUAAUUGCCUUUGUUUUUGCCGCCGCUAUUACUGCUUUUGGUACCAUUGAACUC (--------(((((((.(((((....))))).((((((((....))).))))).)))).))))(((((..((....))..)))))(((((.......))....)))............ ( -31.10) >consensus AUUUUGACAUACGCACCGCACAAUAUUAUGCCCACCACUGUUGGCAGUGGGUGUGUGUGUCAUGGUAAUUGCCUUUGUUUUUGCUGCCGCUAUUGCUGCUUUUGGUACCAUUGAACUC ....((((((((((((((((........)))....(((((....)))))))))))))))))).(((....)))...((((.((.((((((.......))....)))).))..)))).. (-27.21 = -30.10 + 2.89)

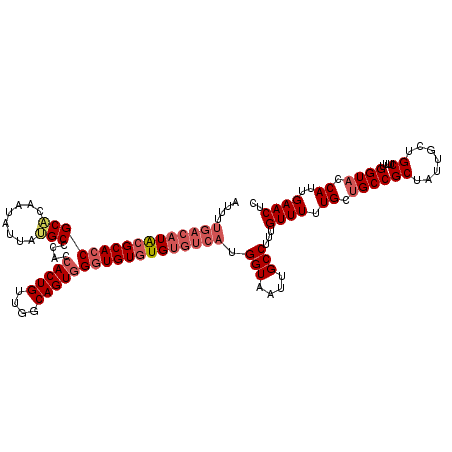
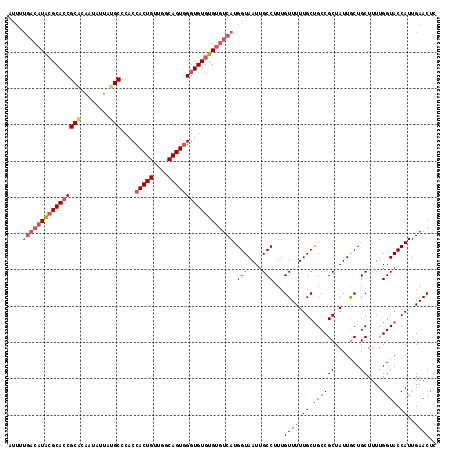
| Location | 19,158,913 – 19,159,031 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -25.86 |
| Energy contribution | -25.87 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19158913 118 - 22407834 GAGUUCAAUGGUACCAAAAGCAGCAAUAGCGGCAGCAAAAACAAAGGCAAUUACCAUGACACACACACCCACUGCCAACAGUGGUGGGCAUAAUAUUGUGCGGUGCGUAUGUCAAAAU .......((((((......((.((....)).)).((..........))...))))))((((.((.((((..(..(((....)))..)(((((....))))))))).)).))))..... ( -34.50) >DroSim_CAF1 39444 118 - 1 GAGUUCAAUGGUACCAAAAGCAGCAAUAGCGGCAGCAAAAACAAAGGCAAUUACCAUGACACACACACCCACUGCCAACAGUGGUGGGCAUAAUAUUGAGCGCUGCGUAUGUCAAAAU .......((((((......((.((....)).)).((..........))...))))))((((.((.(((((((..(.....)..)))))((......)).....)).)).))))..... ( -28.10) >DroYak_CAF1 37762 110 - 1 GAGUUCAAUGGUACCAAAAGCAGUAAUAGCGGCGGCAAAAACAAAGGCAAUUACCAUGACACACACACCAACUGACAACAGUGGUGGGCCCAAUAUUGGGCGGUGCUCA--------U (((.(((.(((((......((.((....)).)).((..........))...))))))))..(((.(((((.(((....)))))))).(((((....))))).)))))).--------. ( -31.60) >consensus GAGUUCAAUGGUACCAAAAGCAGCAAUAGCGGCAGCAAAAACAAAGGCAAUUACCAUGACACACACACCCACUGCCAACAGUGGUGGGCAUAAUAUUGAGCGGUGCGUAUGUCAAAAU ..(((..((((((......((.((....)).)).((..........))...))))))))).(((.((((.((((....)))))))).(((((....))))).)))............. (-25.86 = -25.87 + 0.01)

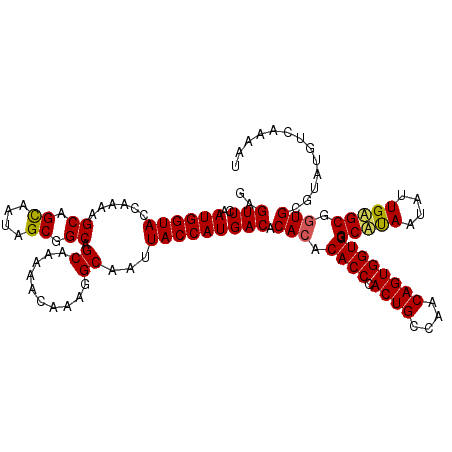
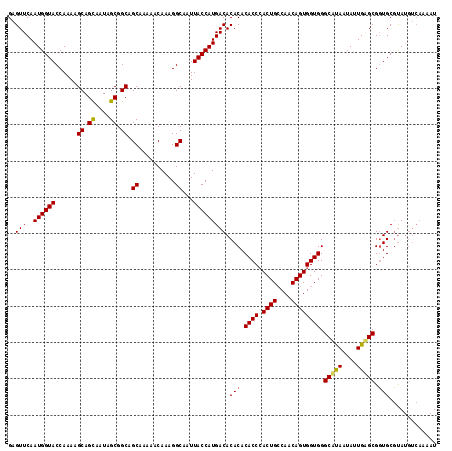
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:18 2006