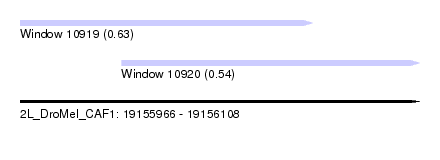
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,155,966 – 19,156,108 |
| Length | 142 |
| Max. P | 0.628533 |

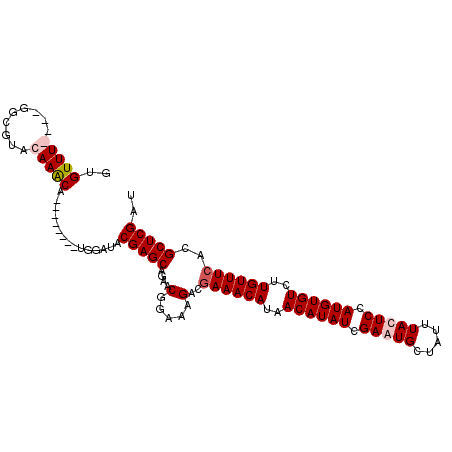
| Location | 19,155,966 – 19,156,070 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -21.55 |
| Energy contribution | -22.55 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19155966 104 + 22407834 GGGUUU----GGCGUACAAGCAUGUGGAUAUGGAUACGAGCAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUACUCCAUGUGUCUUGUUUCACGCUCGAU ..((((----(.....)))))...............(((((.....(......)((((((..((((((.((...........)).))))))..))))))..))))).. ( -25.20) >DroSim_CAF1 36484 104 + 1 GUGUUU----GGCGUACAAGCAUGUGGAUAUGGAUACGAGCAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUACUCCAUGUGUCUUGUUUCACGCUCGAU ((((((----(.....))))))).............(((((.....(......)((((((..((((((.((...........)).))))))..))))))..))))).. ( -27.20) >DroEre_CAF1 35112 100 + 1 GUGUUUGGUUGGUGUACAAACA--------UGGAUACGAGCAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUAUUCCAUGUGUCUUGUUUCACGCUCGAU (((((((.........))))))--------).....(((((.....(......)((((((..((((((.(((((.....))))).))))))..))))))..))))).. ( -30.10) >DroYak_CAF1 35044 97 + 1 GUGUUUGGUUGGCGUACAAACA--------UGGAUACGAGCAGAACGGAAAG---AAACAUAACAUAUCGAAUGCUAUUUAUUCCAUGUGUCUUGUUUCACGCUCGAU (((((((.........))))))--------).....(((((..........(---(((((..((((((.(((((.....))))).))))))..))))))..))))).. ( -26.00) >consensus GUGUUU____GGCGUACAAACA________UGGAUACGAGCAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUACUCCAUGUGUCUUGUUUCACGCUCGAU ..(((((.........)))))...............(((((....(.....)..((((((..((((((.(((((.....))))).))))))..))))))..))))).. (-21.55 = -22.55 + 1.00)

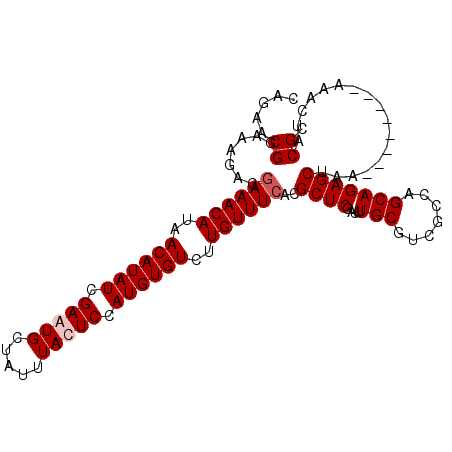
| Location | 19,156,002 – 19,156,108 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -20.62 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19156002 106 + 22407834 CAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUACUCCAUGUGUCUUGUUUCACGCUCGAUUUGCGUCGCCAGCAGAGCUAAGAAAUAAAAAACUACGC ......((...(((((((((..((((((.((...........)).))))))..))))))..((.......))))).))(((...)))................... ( -20.90) >DroSim_CAF1 36520 106 + 1 CAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUACUCCAUGUGUCUUGUUUCACGCUCGAUUUGCGUCGCCAGCAGAGCUAAAAAAUAUAAAACUACGC ......((...(((((((((..((((((.((...........)).))))))..))))))..((.......))))).))(((...)))................... ( -20.90) >DroEre_CAF1 35144 98 + 1 CAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUAUUCCAUGUGUCUUGUUUCACGCUCGAUUUGCGUCGCCAGCAGAGCUAA--------AAACUACGC ......((...(((((((((..((((((.(((((.....))))).))))))..))))))..((.......))))).))(((...)))..--------......... ( -24.60) >DroYak_CAF1 35076 95 + 1 CAGAACGGAAAG---AAACAUAACAUAUCGAAUGCUAUUUAUUCCAUGUGUCUUGUUUCACGCUCGAUUUGCGUCGCCAGCAGAGCUAA--------AAACUACGC .....((....(---(((((..((((((.(((((.....))))).))))))..))))))..((((....(((.......)))))))...--------......)). ( -23.50) >consensus CAGAACGGAAAGACGAAACAUAACAUAUCGAAUGCUAUUUACUCCAUGUGUCUUGUUUCACGCUCGAUUUGCGUCGCCAGCAGAGCUAA________AAACUACGC .....((.......((((((..((((((.(((((.....))))).))))))..))))))..((((....(((.......))))))).................)). (-20.62 = -21.38 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:15 2006