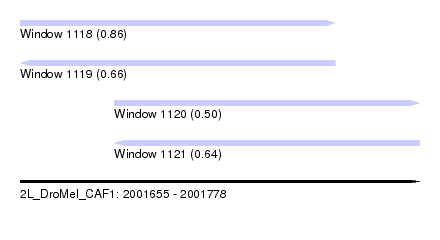
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,001,655 – 2,001,778 |
| Length | 123 |
| Max. P | 0.860007 |

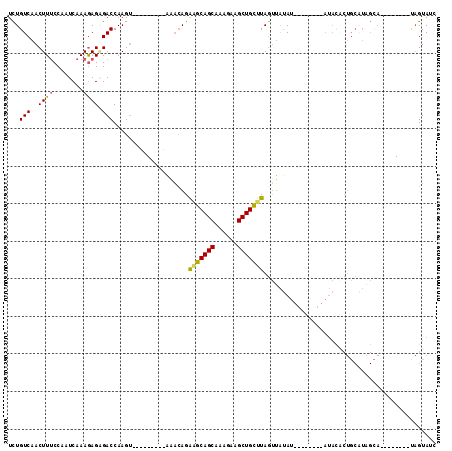
| Location | 2,001,655 – 2,001,752 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.56 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -13.48 |
| Energy contribution | -12.95 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2001655 97 + 22407834 GAUACUA--------UGCUAUGCAGGGUAUAUGUACAUAUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGA ((.((((--------.(.((.((((((((((((....)))))).....(((((......))))))))))).)---------)).)))).))((((..(((....)))..).))) ( -23.70) >DroSec_CAF1 19250 89 + 1 GCUACUA--------UGCUAUGCAGUGUAU--------AUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGA .....((--------(((........))))--------).........(((((......))))).((((((.---------((((.(((.......)))....)))).)))))) ( -20.60) >DroSim_CAF1 16108 89 + 1 GAUACUA--------UGCAAUGCAGUGUAU--------AUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGA .((((..--------(((...)))..))))--------..........(((((......))))).((((((.---------((((.(((.......)))....)))).)))))) ( -21.30) >DroEre_CAF1 16198 104 + 1 GAUACUAUGCCACUAUGCUACGCAGUGUAU--------AUAUAACUAAGCAGCUUCUUUGCUGCUCCU--CCACUUGGCUUACUUGGUCCCUUUUUGAUUGGAAAGUUGACAGA ........((((.(((((........))))--------)........((((((......))))))...--.....)))).......(((.((((((....))))))..)))... ( -22.60) >DroYak_CAF1 20620 103 + 1 GAUACUGUGCCACUAUUCUAUGCAGUAUAU--------AUAUAACUAAGCAGCUUCUUUGCUGCUUCUGUUUACUUGGU---CUUGGUCGCUCUUUGAUUGGAAAGUUGACAGA .(((((((.............)))))))..--------.........((((((......))))))((((((.((((...---.(..((((.....))))..).)))).)))))) ( -27.82) >DroAna_CAF1 15885 74 + 1 UAUAC-------------------------------AUGUCCUGUUAGUCAGCUUCUUUGCUGAUUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGUAAGUUGACAAA .....-------------------------------.((((.....(((((((......)))))))...(((---------(((.((((.......))))))))))..)))).. ( -15.60) >consensus GAUACUA________UGCUAUGCAGUGUAU________AUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU_________ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGA .........................................(((((..(((((......)))))...................(..(((.......)))..)..)))))..... (-13.48 = -12.95 + -0.53)

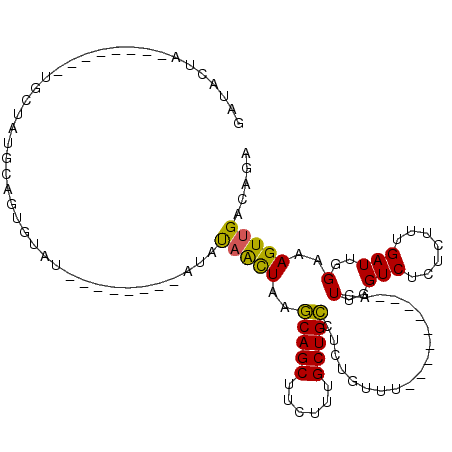
| Location | 2,001,655 – 2,001,752 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.56 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -11.85 |
| Energy contribution | -11.47 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2001655 97 - 22407834 UCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU---------AAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAUAUGUACAUAUACCCUGCAUAGCA--------UAGUAUC (((((..((((.....((.....))....))))---------..)))))((((((......))))))...((((.((((((.........))))))..)--------))).... ( -19.20) >DroSec_CAF1 19250 89 - 1 UCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU---------AAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAU--------AUACACUGCAUAGCA--------UAGUAGC (((((..((((.....((.....))....))))---------..)))))((((((......)))))).........--------.....((((......--------..)))). ( -18.60) >DroSim_CAF1 16108 89 - 1 UCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU---------AAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAU--------AUACACUGCAUUGCA--------UAGUAUC (((((..((((.....((.....))....))))---------..)))))((((((......)))))).........--------((((..(((...)))--------..)))). ( -18.10) >DroEre_CAF1 16198 104 - 1 UCUGUCAACUUUCCAAUCAAAAAGGGACCAAGUAAGCCAAGUGG--AGGAGCAGCAAAGAAGCUGCUUAGUUAUAU--------AUACACUGCGUAGCAUAGUGGCAUAGUAUC .((((..(((((((..........)))..))))..((((.((((--..(((((((......)))))))..))))..--------.(((.....)))......)))))))).... ( -23.00) >DroYak_CAF1 20620 103 - 1 UCUGUCAACUUUCCAAUCAAAGAGCGACCAAG---ACCAAGUAAACAGAAGCAGCAAAGAAGCUGCUUAGUUAUAU--------AUAUACUGCAUAGAAUAGUGGCACAGUAUC .((((..((((((...((.......))....)---)..))))..))))(((((((......)))))))........--------..(((((((((......)))...)))))). ( -18.40) >DroAna_CAF1 15885 74 - 1 UUUGUCAACUUACCAAUCAAAGAGAGACCAAGU---------AAACAGAAUCAGCAAAGAAGCUGACUAACAGGACAU-------------------------------GUAUA ..((((.((((.....((.....))....))))---------........(((((......))))).......)))).-------------------------------..... ( -11.30) >consensus UCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU_________AAACAGAAGCAGCAAAGAAGCUGCUUAGUUAUAU________AUACACUGCAUAGCA________UAGUAUC ...(((..((((........)))).)))....................(((((((......))))))).............................................. (-11.85 = -11.47 + -0.39)

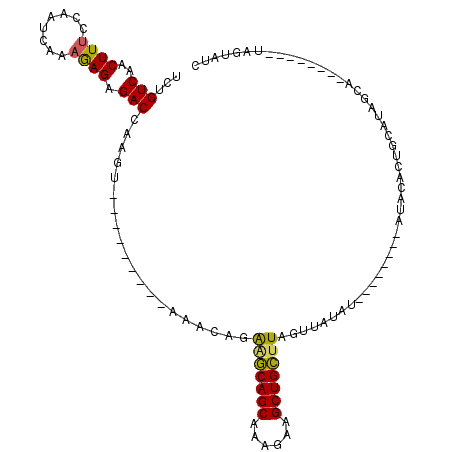
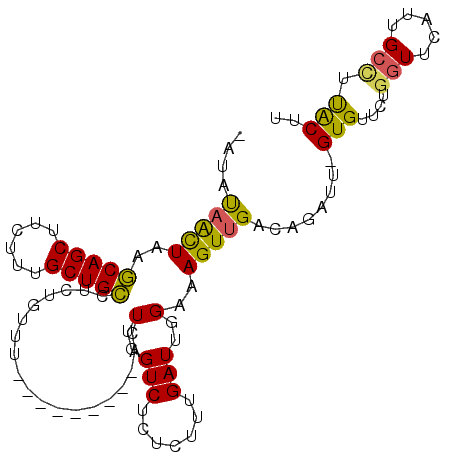
| Location | 2,001,684 – 2,001,778 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -14.78 |
| Energy contribution | -14.20 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2001684 94 + 22407834 UAUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGAUU-GUGUUCCGGUUCAUUGCCUUACUU .((((((....(((((......))))).((((((.---------((((.(((.......)))....)))).))))))))-))))...(((.....)))...... ( -22.40) >DroSec_CAF1 19272 93 + 1 -AUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGAUU-GUGUUCUGGUUCAUUGCUUCACUU -..........(((((......))))).((((((.---------((((.(((.......)))....)))).))))))..-(((....(((.....))).))).. ( -21.40) >DroSim_CAF1 16130 93 + 1 -AUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGAUU-GUGUUCUGGUUCAUUGCCUUACUU -((((((....(((((......))))).((((((.---------((((.(((.......)))....)))).))))))))-))))...(((.....)))...... ( -21.90) >DroEre_CAF1 16228 100 + 1 -AUAUAACUAAGCAGCUUCUUUGCUGCUCCU--CCACUUGGCUUACUUGGUCCCUUUUUGAUUGGAAAGUUGACAGAUU-GUGUUCUGGUUCAUUGCCUUACUU -.........((((((......))))))...--......(((...............(..(((....)))..)((((..-....)))).......)))...... ( -20.60) >DroYak_CAF1 20650 100 + 1 -AUAUAACUAAGCAGCUUCUUUGCUGCUUCUGUUUACUUGGU---CUUGGUCGCUCUUUGAUUGGAAAGUUGACAGAUUCGUGUUCUGGUUCAUUGCCUUACUU -.........((((((......))))))((((((.((((...---.(..((((.....))))..).)))).))))))...(((....(((.....))).))).. ( -25.70) >DroAna_CAF1 15891 94 + 1 UGUCCUGUUAGUCAGCUUCUUUGCUGAUUCUGUUU---------ACUUGGUCUCUCUUUGAUUGGUAAGUUGACAAAUG-GUGUGUUAGUUCAUCGAGUUGCCU .........(((((((......)))))))......---------....(((..(((.(..(((....)))..).....(-(((........)))))))..))). ( -19.50) >consensus _AUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUU_________ACUUGGUCUCUCUUUGAUUGGAAAGUUGACAGAUU_GUGUUCUGGUUCAUUGCCUUACUU ....(((((..(((((......)))))...................(..(((.......)))..)..)))))........(((....(((.....))).))).. (-14.78 = -14.20 + -0.58)

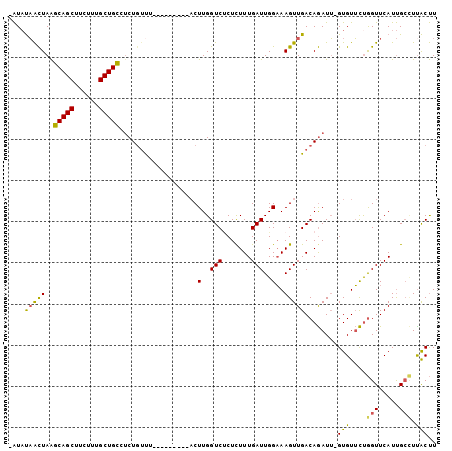
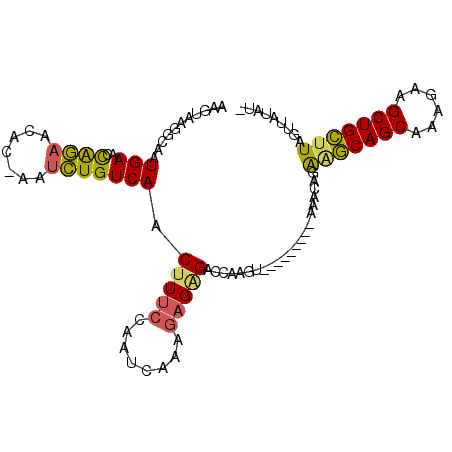
| Location | 2,001,684 – 2,001,778 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -19.07 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.35 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2001684 94 - 22407834 AAGUAAGGCAAUGAACCGGAACAC-AAUCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU---------AAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAUA ......((...(((..((((....-..))))))).(((((........))))).))..((---------((....(((((((......)))))))..))))... ( -19.20) >DroSec_CAF1 19272 93 - 1 AAGUGAAGCAAUGAACCAGAACAC-AAUCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU---------AAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAU- ..((...((..(((..((((....-..))))))).(((((........))))).....))---------..))..(((((((......)))))))........- ( -18.40) >DroSim_CAF1 16130 93 - 1 AAGUAAGGCAAUGAACCAGAACAC-AAUCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU---------AAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAU- ......((...(((..((((....-..))))))).(((((........))))).))..((---------((....(((((((......)))))))..))))..- ( -19.90) >DroEre_CAF1 16228 100 - 1 AAGUAAGGCAAUGAACCAGAACAC-AAUCUGUCAACUUUCCAAUCAAAAAGGGACCAAGUAAGCCAAGUGG--AGGAGCAGCAAAGAAGCUGCUUAGUUAUAU- ......(((..(((..((((....-..)))))))(((((((..........)))..))))..)))..((((--..(((((((......)))))))..))))..- ( -25.10) >DroYak_CAF1 20650 100 - 1 AAGUAAGGCAAUGAACCAGAACACGAAUCUGUCAACUUUCCAAUCAAAGAGCGACCAAG---ACCAAGUAAACAGAAGCAGCAAAGAAGCUGCUUAGUUAUAU- .......((..(((..((((.......))))))).((((......)))).)).......---.....((((....(((((((......)))))))..))))..- ( -17.40) >DroAna_CAF1 15891 94 - 1 AGGCAACUCGAUGAACUAACACAC-CAUUUGUCAACUUACCAAUCAAAGAGAGACCAAGU---------AAACAGAAUCAGCAAAGAAGCUGACUAACAGGACA ((....))...............(-(.(((((..((((.....((.....))....))))---------..))))).(((((......)))))......))... ( -14.40) >consensus AAGUAAGGCAAUGAACCAGAACAC_AAUCUGUCAACUUUCCAAUCAAAGAGAGACCAAGU_________AAACAGAAGCAGCAAAGAAGCUGCUUAGUUAUAU_ ...........(((..((((.......))))))).(((((........)))))......................(((((((......)))))))......... (-13.76 = -13.35 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:52 2006