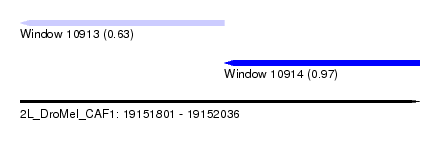
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,151,801 – 19,152,036 |
| Length | 235 |
| Max. P | 0.969442 |

| Location | 19,151,801 – 19,151,921 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
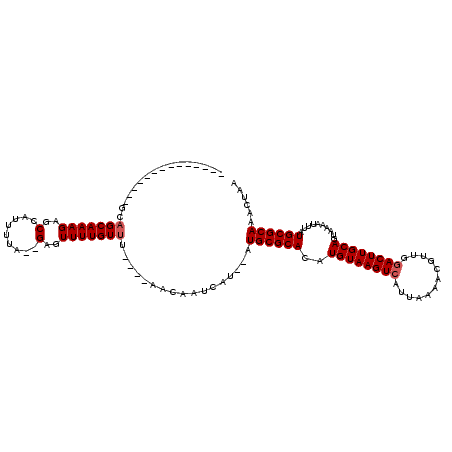
>2L_DroMel_CAF1 19151801 120 - 22407834 AUAGAAUUGUACUAGCAGCAAAGAGCCAUUUUAAAGAGUUUUGUUUAACAAACAAUCAUGUGUGCGCACAUGUAAGUCAUAAAAACGUUGGACUUGCAGUAAAAUUUAUGCGCAAACUAU ...((.((((......(((((((..(.........)..)))))))......))))))..((.((((((..((((((((.(((.....)))))))))))..........)))))).))... ( -22.60) >DroEre_CAF1 31100 98 - 1 --------------GCAGCAAAGAGCCAUUUUA--GAUUUUUGUUA----AACAAUCAU--AUGCGCACAUGUAAGUCAUUAAAACGGUGGACUUGCAGUAAAAUUUAUGCGCAAACUAA --------------..((((((((..(......--).)))))))).----.........--.((((((..(((((((((((.....))).))))))))..........))))))...... ( -20.20) >DroYak_CAF1 31250 98 - 1 --------------AGCGCAAAGAGCCAUUUUA--GAGUUUUGUUU----AACAAUAAU--AUGCGCACAUGUAAGUCAUUAAAACGUUGAACUUGCAGUAAAAUUUAUGCGCAAACUAA --------------.(((((.((((((((.(((--..(((......----)))..))).--))).))((.(((((((((.........)).)))))))))....))).)))))....... ( -23.00) >consensus ______________GCAGCAAAGAGCCAUUUUA__GAGUUUUGUUU____AACAAUCAU__AUGCGCACAUGUAAGUCAUUAAAACGUUGGACUUGCAGUAAAAUUUAUGCGCAAACUAA ................(((((((..(.........)..))))))).................((((((..((((((((............))))))))..........))))))...... (-15.50 = -16.17 + 0.67)

| Location | 19,151,921 – 19,152,036 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
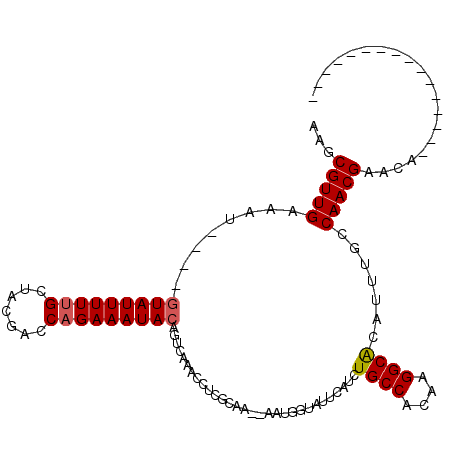
>2L_DroMel_CAF1 19151921 115 - 22407834 AAGCGUUGAAAUACAUAUAUUUUUGCUACGACCAGAAAUACAGUCAAGCCUCGCAAAAAAUGCUAUUCAUCUGCCACAAGGCGCAUUUGACAACGAACCACGGAACCAUUCAAAC .....(((((.......((((((((.......))))))))..(((((((...(((.....))).........(((....)))))..))))).................))))).. ( -18.20) >DroEre_CAF1 31198 93 - 1 AAGCGUUGAAAU----GUAUUUUUGCUACGACCUGAAAUACAGUCAAACCUCGCAA--AAUCGUAUUCAUCUGCCACAAGGCACAUUUGCCAACGAAAU---------------- ..(((((((..(----(((((((...........)))))))).))))....)))..--..((((...((..((((....))))....))...))))...---------------- ( -17.80) >DroYak_CAF1 31348 92 - 1 AAGCGUUGAAAU----GUAUUUUUGCUACGAGCAGAAAUACAGUCAAACCUCGCAA---AUGGUAUUCAUCUGCCACAAGGCACAUUUACCAACGAACA---------------- ..(((((((..(----((((((((((.....))))))))))).))))....)))..---.(((((...((.((((....)))).)).))))).......---------------- ( -28.00) >consensus AAGCGUUGAAAU____GUAUUUUUGCUACGACCAGAAAUACAGUCAAACCUCGCAA__AAUGGUAUUCAUCUGCCACAAGGCACAUUUGCCAACGAACA________________ ...(((((........(((((((((.......)))))))))..............................((((....)))).......))))).................... (-14.99 = -15.43 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:09 2006