
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,138,842 – 19,138,944 |
| Length | 102 |
| Max. P | 0.798324 |

| Location | 19,138,842 – 19,138,944 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.75 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
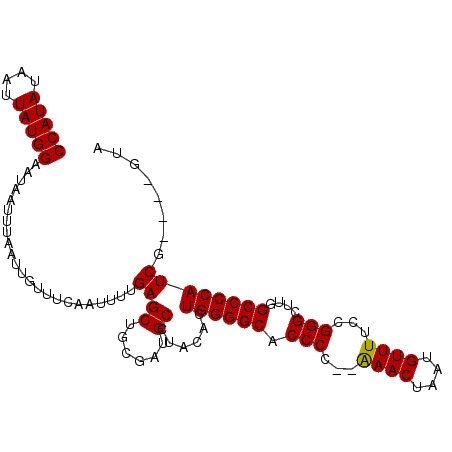
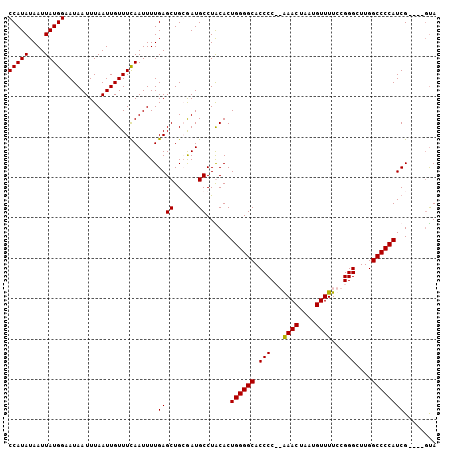
>2L_DroMel_CAF1 19138842 102 + 22407834 CCAUAUAAUUAUGGAAUAAUUUAAUUGUUUCAAUUUUGAGCUGCGAUGCCUACACUGGGGCACCCCCGAAACUAAUGUUUUCCGGGCUUGGCCCCAUCG----UUA (((((....))))).........(((((((((....))))..)))))........((((((...((((((((....))))..))))....))))))...----... ( -27.30) >DroSim_CAF1 26574 102 + 1 CCAUAUAAUUAUGGAAUAAUUUAAUUGUUUCAAUUUUGAGCUGCGAUGCCUGCACUGGGGCACCCCCGAAACUAAUGUUUUCCGGGCUUGGCCCCAUCG----GCA (((((....))))).........(((((((((....))))..)))))(((.....((((((...((((((((....))))..))))....))))))..)----)). ( -31.10) >DroEre_CAF1 25208 100 + 1 CCAUAUAAUUAUGGAAUAAUUUAAUUGUUUCAAUUUUGAGCUGGAAUGCCUACACUGGGGCACCCC--GAACUAAUGUUUUCCGGGCUUGGCCCCAUCG----GUA (((..(((...(((((((((...)))))))))...)))...)))..((((.....((((((..(((--((((....)))...))))....))))))..)----))) ( -26.50) >DroYak_CAF1 25393 104 + 1 CCAUAUAAUUAUGGAAUAAUUUAAUUGUUUUAAUUUUGAGCUGGGAUGCCUGCACUGGGGCACCCC--GAACUAAUGUUUUCCGGGCUUGGCCCCAUCGCGUCGUA (((((....)))))............((((.......))))...(((((......((((((..(((--((((....)))...))))....))))))..)))))... ( -28.50) >consensus CCAUAUAAUUAUGGAAUAAUUUAAUUGUUUCAAUUUUGAGCUGCGAUGCCUACACUGGGGCACCCC__AAACUAAUGUUUUCCGGGCUUGGCCCCAUCG____GUA (((((....))))).......................((((......))......((((((.(((...((((....))))...)))....))))))))........ (-24.00 = -23.75 + -0.25)

| Location | 19,138,842 – 19,138,944 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -21.89 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
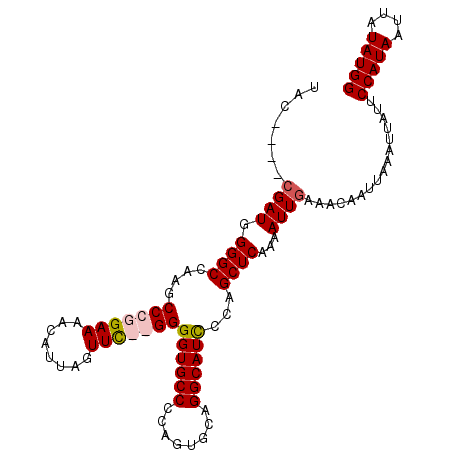
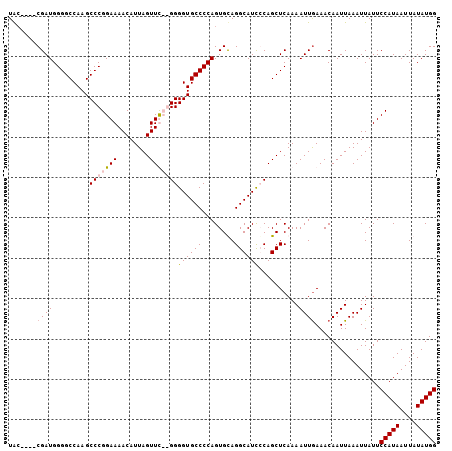
>2L_DroMel_CAF1 19138842 102 - 22407834 UAA----CGAUGGGGCCAAGCCCGGAAAACAUUAGUUUCGGGGGUGCCCCAGUGUAGGCAUCGCAGCUCAAAAUUGAAACAAUUAAAUUAUUCCAUAAUUAUAUGG ...----((((.((((....(((((((........)))))))((((((........))))))...))))...))))................(((((....))))) ( -27.90) >DroSim_CAF1 26574 102 - 1 UGC----CGAUGGGGCCAAGCCCGGAAAACAUUAGUUUCGGGGGUGCCCCAGUGCAGGCAUCGCAGCUCAAAAUUGAAACAAUUAAAUUAUUCCAUAAUUAUAUGG (((----(..(((((((...(((((((........))))))).).)))))).....))))................................(((((....))))) ( -30.00) >DroEre_CAF1 25208 100 - 1 UAC----CGAUGGGGCCAAGCCCGGAAAACAUUAGUUC--GGGGUGCCCCAGUGUAGGCAUUCCAGCUCAAAAUUGAAACAAUUAAAUUAUUCCAUAAUUAUAUGG (((----...((((((...((((.(((........)))--.))))))))))..)))(((......)))........................(((((....))))) ( -26.20) >DroYak_CAF1 25393 104 - 1 UACGACGCGAUGGGGCCAAGCCCGGAAAACAUUAGUUC--GGGGUGCCCCAGUGCAGGCAUCCCAGCUCAAAAUUAAAACAAUUAAAUUAUUCCAUAAUUAUAUGG ......(((.((((((...((((.(((........)))--.)))))))))).))).(((......)))........................(((((....))))) ( -29.40) >consensus UAC____CGAUGGGGCCAAGCCCGGAAAACAUUAGUUC__GGGGUGCCCCAGUGCAGGCAUCCCAGCUCAAAAUUGAAACAAUUAAAUUAUUCCAUAAUUAUAUGG .......((((.((((....(((((((........)))))))((((((........))))))...))))...))))................(((((....))))) (-21.89 = -22.70 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:05 2006